6C4X
 
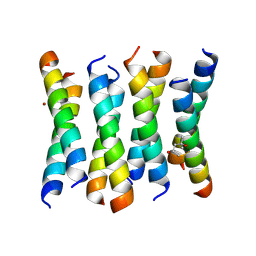 | | Cross-alpha Amyloid-like Structure alphaAmmem | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ZINC ION, cross-alpha amyloid-like membrane peptide alpha-AmMEM | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C50
 
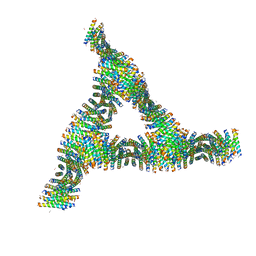 | | Cross-alpha Amyloid-like Structure alphaAmS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmS, FORMIC ACID | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C4Z
 
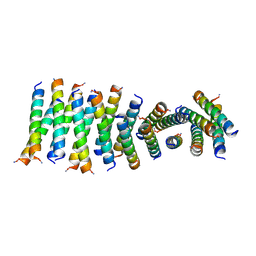 | |
6C4Y
 
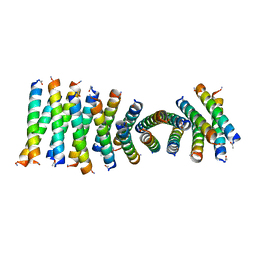 | | Cross-alpha Amyloid-like Structure alphaAmG | | Descriptor: | Cross-alpha Amyloid-like Structure alphaAmG | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C52
 
 | | Cross-alpha Amyloid-like Structure alphaTet | | Descriptor: | Cross-alpha Amyloid-like Structure alphaTet, GLYCEROL | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C51
 
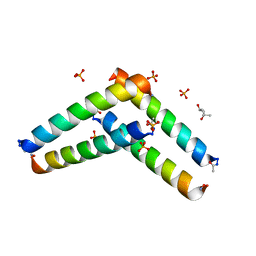 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6D02
 
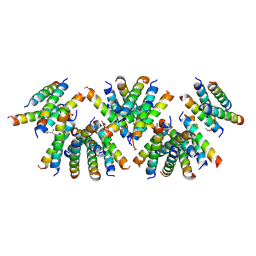 | | Cross-alpha Amyloid-like Structure alphaAmL, 2nd form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.-Q, Liu, L, Degrado, W.F. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6O3N
 
 | |
5WLL
 
 | |
5WLJ
 
 | |
5WLM
 
 | |
5WLK
 
 | |
7YPG
 
 | |
5HKN
 
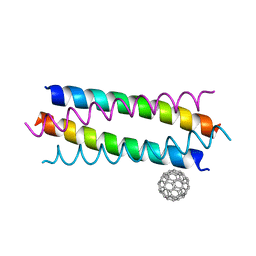 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5HKR
 
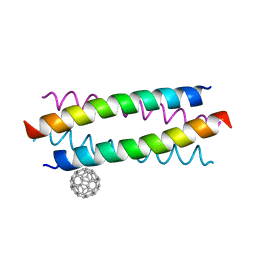 | | Crystal structure of de novo designed fullerene organising protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Acharya, R, Kim, Y.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5TGY
 
 | | NMR structure of holo-PS1 | | Descriptor: | PS1, [5,10,15,20-tetrakis(trifluoromethyl)porphyrinato(2-)-kappa~4~N~21~,N~22~,N~23~,N~24~]zinc | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
5TGW
 
 | | NMR structure of apo-PS1 | | Descriptor: | PS1 | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
7V8O
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of P21221 | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexanone Monooxygenase from Thermocrispum municipale, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
7V8S
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of P1211 | | Descriptor: | Cyclohexanone Monooxygenase from Thermocrispum municipale, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
7V8R
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of C2221 | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexanone Monooxygenase from Thermocrispum municipale, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
