3CUP
 
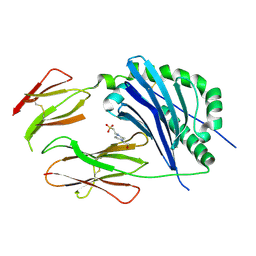 | | Crystal structure of the MHC class II molecule I-Ag7 in complex with the peptide GAD221-235 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, H-2 class II histocompatibility antigen, ... | | Authors: | Corper, A.L, Yoshida, K, Teyton, L, Wilson, I.A. | | Deposit date: | 2008-04-16 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of the MHC class II molecule I-Ag7 in complex with the peptide GAD221-235
To be Published
|
|
3MBE
 
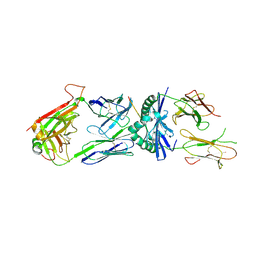 | | TCR 21.30 in complex with MHC class II I-Ag7HEL(11-27) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II H2-IAg7 ALPHA CHAIN, ... | | Authors: | Corper, A.L, Yoshida, K, Teyton, L, Wilson, I.A. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | The diabetogenic mouse MHC class II molecule I-Ag7 is endowed with a switch that modulates TCR affinity.
J.Clin.Invest., 120, 2010
|
|
4YVI
 
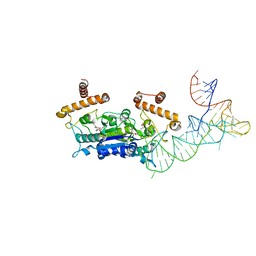 | |
4YVJ
 
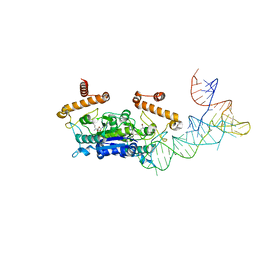 | |
4YVK
 
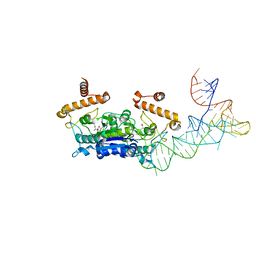 | |
4YVH
 
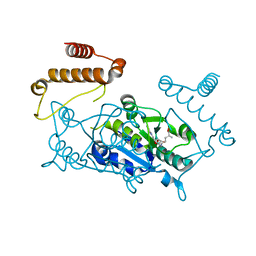 | |
3AXZ
 
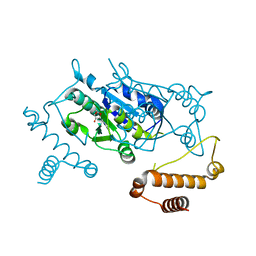 | | Crystal structure of Haemophilus influenzae TrmD in complex with adenosine | | Descriptor: | ADENOSINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Yoshida, K, Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
3RQW
 
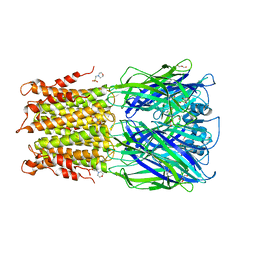 | | Crystal structure of acetylcholine bound to a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYLCHOLINE, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, ... | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
3RQU
 
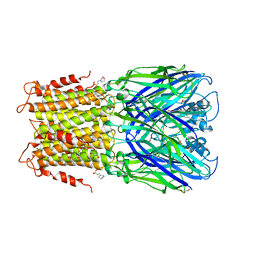 | | Crystal structure of a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, GLYCEROL | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
1MKN
 
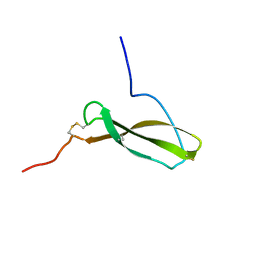 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1MKC
 
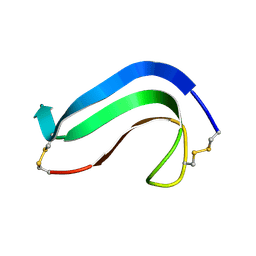 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
3PBP
 
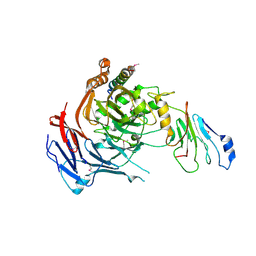 | |
8ABX
 
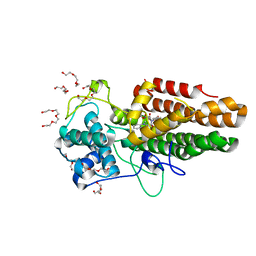 | | Crystal structure of IDO1 in complex with Apoxidole-1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, O1-tert-butyl O2-ethyl O5-methyl (E,5R)-5-(1-methylindol-2-yl)-5-[(4-methylphenyl)sulfonylamino]pent-2-ene-1,2,5-tricarboxylate, O2-tert-butyl O3-ethyl O6-methyl (2S,6R)-6-(1-methylindol-2-yl)-2,5-dihydro-1H-pyridine-2,3,6-tricarboxylate, ... | | Authors: | Dotsch, L, Ziegler, S, Waldmann, H, Gasper, R. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Pseudo-Natural Product Type IV IDO1 Inhibitor Chemotype.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7NS0
 
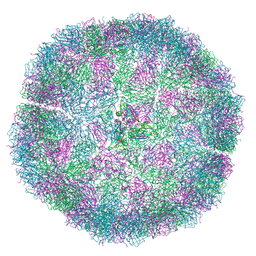 | | Bacilladnavirus capsid structure | | Descriptor: | Capsid protein VP2 | | Authors: | Munke, A, Okamoto, K. | | Deposit date: | 2021-03-05 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses.
Mbio, 13, 2022
|
|
3VJK
 
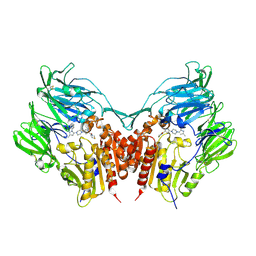 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
6HUS
 
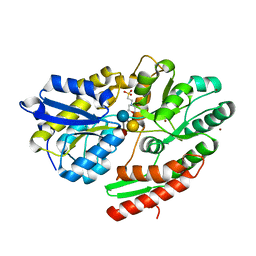 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
3VJL
 
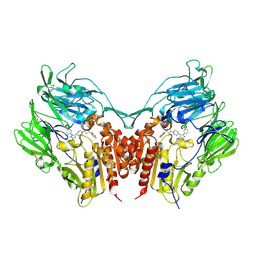 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
7D7P
 
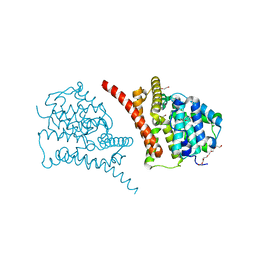 | | Crystal structure of the phosphodiesterase domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
6HUR
 
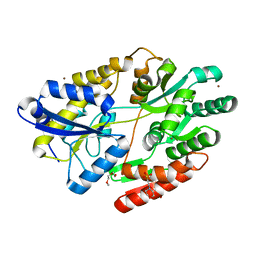 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 2'-fucosyllactose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Katayama, T, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
7M41
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-methyl-5-oxo-5,6-dihydro-[1,2,4]traizolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)-1H-imidazole-2-sulfonamide (compound 38) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}-1H-imidazole-2-sulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
7M3Y
 
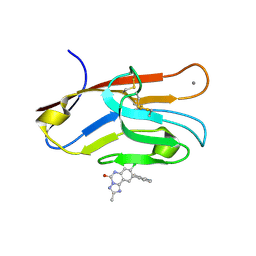 | | Structure of TIM-3 in complex with 8-chloro-2-methyl-9-(3-mehtylpyridin-4-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5(6H)-one (compound 22) | | Descriptor: | (4R,10aP)-8-chloro-2-methyl-9-(3-methylpyridin-4-yl)[1,2,4]triazolo[1,5-c]quinazolin-5(6H)-one, CALCIUM ION, Hepatitis A virus cellular receptor 2 | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
7M3Z
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-mehtyl-5-oxo-5,6-dihydro-[1,2,4]triazolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)methanesulfonamdide (compound 35) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}methanesulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
7CJ3
 
 | | Crystal structure of the transmembrane domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
7D7Q
 
 | | Crystal structure of the transmembrane domain and linker region of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
7CQY
 
 | |
