8BVE
 
 | | MoeA2 from Corynebacterium glutamicum | | Descriptor: | CITRIC ACID, Molybdopterin molybdenumtransferase, SODIUM ION | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
8BVF
 
 | | MoeA2 from Corynebacterium glutamicum in complex with FtsZ-CTD | | Descriptor: | Cell division protein FtsZ, Molybdopterin molybdenumtransferase, SODIUM ION, ... | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
8AUC
 
 | | Structure of peptidoglycan hydrolase Cg1735 from Corynebacterium glutamicum, trigonal crystal form | | Descriptor: | Cell wall-associated hydrolases (Invasion-associated proteins), TETRACHLOROPLATINATE(II) | | Authors: | Gaday, Q, Wehenkel, A.M, Alzari, P.M, Legrand, P. | | Deposit date: | 2022-08-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | FtsEX-independent control of RipA-mediated cell separation in Corynebacteriales.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8AUD
 
 | |
8AU6
 
 | |
7AL2
 
 | |
7AL1
 
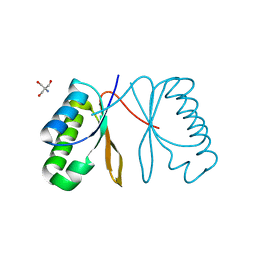 | | Cell division protein SepF from Methanobrevibacter smithii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein SepF | | Authors: | Sogues, A, wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SepF is the FtsZ anchor in archaea, with features of an ancestral cell division system.
Nat Commun, 12, 2021
|
|
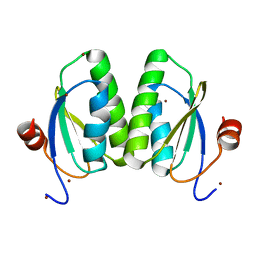
6SCP
 
 | |
6SAT
 
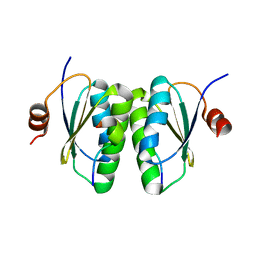 | |
6SCQ
 
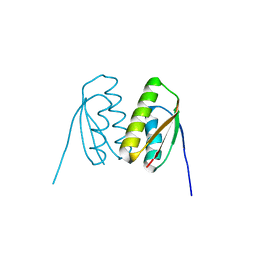 | |
6SCS
 
 | |
2Y0P
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate and acetyl-CoA | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2XT6
 
 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2YIC
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2YID
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2XTA
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with acetyl-CoA (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
8B7F
 
 | | Nuclease from C. glutamicum | | Descriptor: | Ubiquitin-like protein SMT3,MksG | | Authors: | Wehenkel, A, Ben Assaya, M, Haouz, A. | | Deposit date: | 2022-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | The MksG nuclease is the executing part of the bacterial plasmid defense system MksBEFG.
Nucleic Acids Res., 51, 2023
|
|
9GK2
 
 | |
8COY
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) in complex with inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CP0
 
 | | Structure of the catalytic domain of P. vivax Sub1 (trigonal crystal form) | | Descriptor: | CALCIUM ION, subtilisin | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8COZ
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
