9EWX
 
 | | Cryo-EM structure of the Pseudomonas aeruginosa PAO1 Type IV pilus | | Descriptor: | Pilin | | Authors: | Ochner, H, Boehning, J, Wang, Z, Tarafder, A, Caspy, I, Bharat, T.A.M. | | Deposit date: | 2024-04-05 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of the Pseudomonas aeruginosa PAO1 Type-IV pilus
To Be Published
|
|
8CH5
 
 | |
4UF9
 
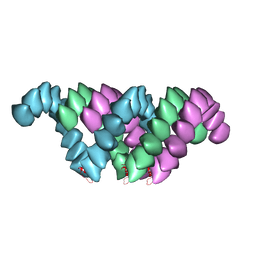 | | Electron cryo-microscopy structure of PB1-p62 type T filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
4UF8
 
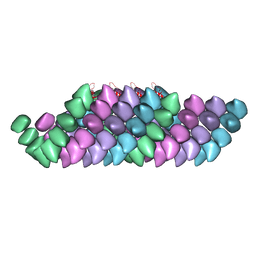 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
8SYL
 
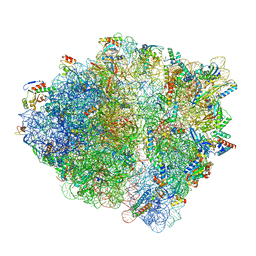 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with amikacin, mRNA, and A-, P-, and E-site tRNAs | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Seely, S.M, Gagnon, M.G. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome.
Nat Commun, 14, 2023
|
|
8EV7
 
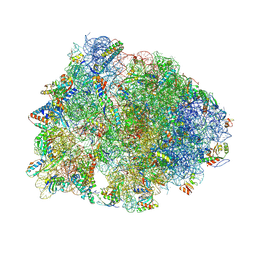 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with kanamycin, mRNA, and A-, P-, and E-site tRNAs | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seely, S.M, Gagnon, M.G. | | Deposit date: | 2022-10-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome.
Nat Commun, 14, 2023
|
|
8EV6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with amikacin, mRNA, and A-, P-, and E-site tRNAs | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Seely, S.M, Gagnon, M.G. | | Deposit date: | 2022-10-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.946 Å) | | Cite: | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome.
Nat Commun, 14, 2023
|
|
