1UWO
 
 | |
3KCP
 
 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
4UAP
 
 | | X-ray structure of GH31 CBM32-2 bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Grondin, J.M, Abe, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
4LKS
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Kirlin, A.C, Furness, H.S, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4LPL
 
 | | Structure of CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Heather, F.S, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4LQR
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4P5Y
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, Glycosyl hydrolase, ... | | Authors: | Grondin, J.M, Allingham, J.S, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-03-04 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
2B59
 
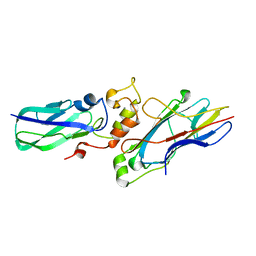 | |
2LS6
 
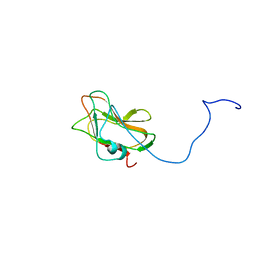 | | Solution NMR Structure of a Non-canonical galactose-binding CBM32 from Clostridium perfringens | | Descriptor: | Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Chitayat, S, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2012-04-20 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An unusual mode of galactose recognition by a family 32 carbohydrate-binding module.
J.Mol.Biol., 426, 2014
|
|
1E88
 
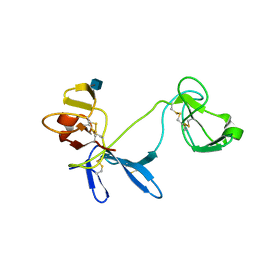 | | Solution structure of 6F11F22F2, a compact three-module fragment of the gelatin-binding domain of human fibronectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIBRONECTIN | | Authors: | Pickford, A.R, Smith, S.P, Staunton, D, Boyd, J, Campbell, I.D. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-09 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The Hairpin Structure of the (6)F1(1)F2(2)F2 Fragment from Human Fibronectin Enhances Gelatin Binding
Embo J., 20, 2001
|
|
1E8B
 
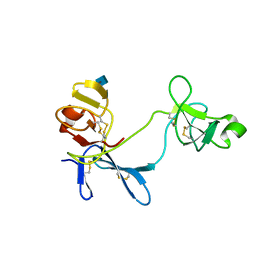 | | Solution structure of 6F11F22F2, a compact three-module fragment of the gelatin-binding domain of human fibronectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIBRONECTIN | | Authors: | Pickford, A.R, Smith, S.P, Staunton, D, Boyd, J, Campbell, I.D. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The Hairpin Structure of the (6)F1(1)F2(2)F2 Fragment from Human Fibronectin Enhances Gelatin Binding
Embo J., 20, 2001
|
|
5SVH
 
 | |
5IBW
 
 | | Complex of MlcC bound to the tandem IQ motif of MyoC | | Descriptor: | Calcium-binding EF-hand domain-containing protein, Myosin IC heavy chain, SODIUM ION | | Authors: | Langelaan, D.N, Smith, S.P. | | Deposit date: | 2016-02-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2FEB
 
 | | NMR Solution Structure, Dynamics and Binding Properties of the Kringle IV Type 8 module of apolipoprotein(a) | | Descriptor: | Apolipoprotein(a) | | Authors: | Chitayat, S, Kanelis, V, Koschinsky, M.L, Smith, S.P. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance (NMR) solution structure, dynamics, and binding properties of the kringle IV type 8 module of apolipoprotein(a).
Biochemistry, 46, 2007
|
|
2JNK
 
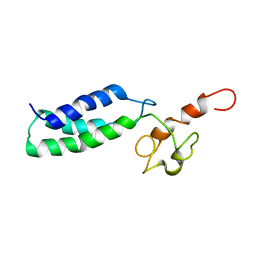 | |
2M1U
 
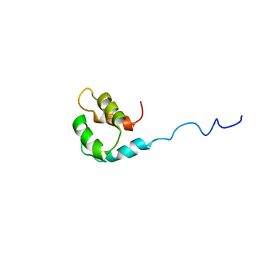 | | Solution structure of the small dictyostelium discoideium myosin light chain mlcb provides insights into iq-motif recognition of class i myosin myo1b | | Descriptor: | myosin light chain MlcB | | Authors: | Liburd, J, Chitayat, S, Crawley, S.W, Denis, C.M, Cote, G.P, Smith, S.P. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the small Dictyostelium discoideum myosin light chain MlcB provides insights into MyoB IQ motif recognition.
J.Biol.Chem., 289, 2014
|
|
2M8U
 
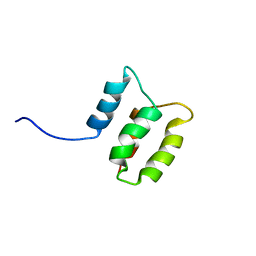 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2O4E
 
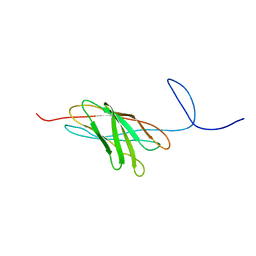 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
2MH0
 
 | |
2OZN
 
 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2KWF
 
 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | Descriptor: | CREB-binding protein, Transcription factor 4 | | Authors: | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | Deposit date: | 2010-04-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
2VO8
 
 | | Cohesin module from Clostridium perfringens ATCC13124 family 33 glycoside hydrolase. | | Descriptor: | EXO-ALPHA-SIALIDASE | | Authors: | Gregg, K, Adams, J.J, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Clostridium Perfringens Toxin Complex Formation.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
