6FZK
 
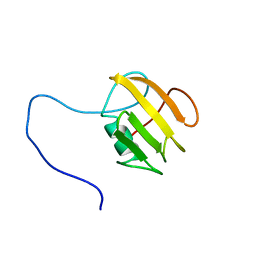 | | NMR structure of UB2H, regulatory domain of PBP1b from E. coli | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Egan, A.J.F, Vollmer, W. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
6G5R
 
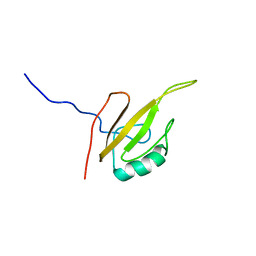 | |
6G5S
 
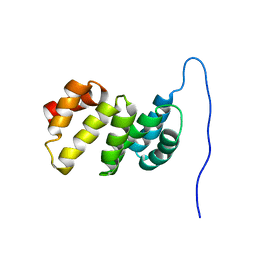 | | Solution structure of the TPR domain of the cell division coordinator, CpoB | | Descriptor: | Cell division coordinator CpoB | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Vollmer, W, Egan, A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
1GH1
 
 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
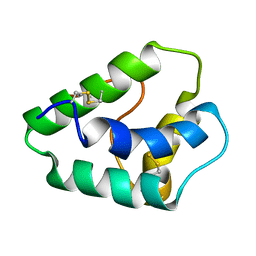
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | Descriptor: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | Deposit date: | 2020-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
4JL0
 
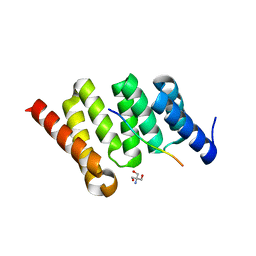 | | Crystal structure of PcrH in complex with the chaperone binding region of PopB | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PopB, Regulatory protein PcrH | | Authors: | Discola, K.F, Forster, A, Simorre, J.P, Attree, I, Dessen, A, Job, V. | | Deposit date: | 2013-03-12 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Membrane and Chaperone Recognition by the Major Translocator Protein PopB of the Type III Secretion System of Pseudomonas aeruginosa.
J.Biol.Chem., 289, 2014
|
|
6ZTG
 
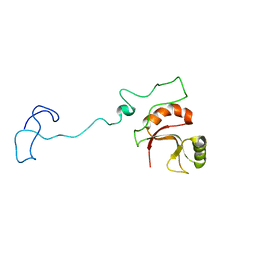 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
2B1W
 
 | |
1I5V
 
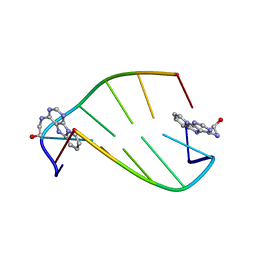 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
7R27
 
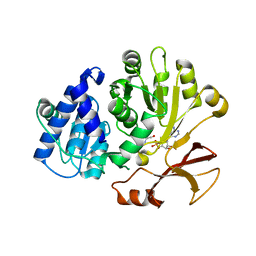 | | Crystal structure of the L. plantarum D-alanine ligase DltA | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-ALANINE, D-alanine--D-alanyl carrier protein ligase | | Authors: | Nikolopoulos, N, Ravaud, S, Simorre, J.P, Grangeasse, C. | | Deposit date: | 2022-02-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | DltC acts as an interaction hub for AcpS, DltA and DltB in the teichoic acid D-alanylation pathway of Lactiplantibacillus plantarum.
Sci Rep, 12, 2022
|
|
7R49
 
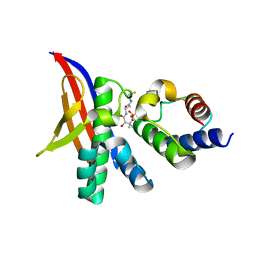 | | Crystal structure of the L. plantarum acyl carrier protein synthase (AcpS)in complex with D-alanyl carrier protein (DltC1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4'-PHOSPHOPANTETHEINE, D-alanyl carrier protein 1, ... | | Authors: | Nikolopoulos, N, Ravaud, S, Simorre, J.P, Grangeasse, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | DltC acts as an interaction hub for AcpS, DltA and DltB in the teichoic acid D-alanylation pathway of Lactiplantibacillus plantarum.
Sci Rep, 12, 2022
|
|
1P6R
 
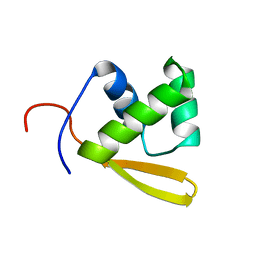 | | Solution structure of the DNA binding domain of the repressor BlaI. | | Descriptor: | Penicillinase repressor | | Authors: | Melckebeke, H.V, Vreuls, C, Gans, P, Llabres, G, Filee, P, Joris, B, Simorre, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structural study of BlaI: implications for the repression of genes involved in beta-lactam antibiotic resistance.
J.Mol.Biol., 333, 2003
|
|
2UWJ
 
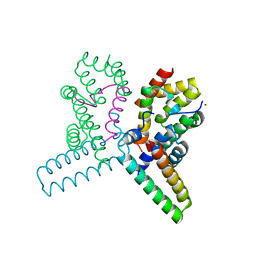 | | Structure of the heterotrimeric complex which regulates type III secretion needle formation | | Descriptor: | NICKEL (II) ION, TYPE III EXPORT PROTEIN PSCE, TYPE III EXPORT PROTEIN PSCF, ... | | Authors: | Quinaud, M, Ple, S, Job, V, Contreras-Martel, C, Simorre, J.P, Attree, I, Dessen, A. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the heterotrimeric complex that regulates type III secretion needle formation.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
6EHZ
 
 | |
3ZGP
 
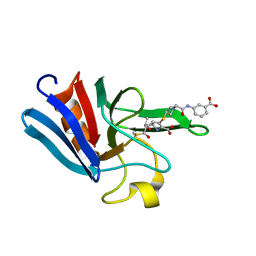 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZG4
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
2MII
 
 | | NMR structure of E. coli LpoB | | Descriptor: | Penicillin-binding protein activator LpoB | | Authors: | Jean, N.L, Egan, A.J.F, Koumoutsi, A, Bougault, C.M, Typas, A, Vollmer, W, Simorre, J.P. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1EHT
 
 | |
6DZ8
 
 | |
2NDA
 
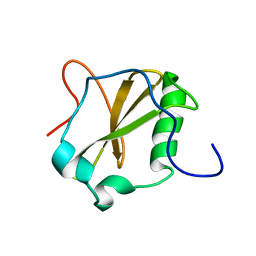 | | Solution structure of MapZ extracellular domain second subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2ND9
 
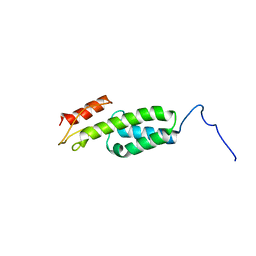 | | Solution structure of MapZ extracellular domain first subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2GMO
 
 | | NMR-structure of an independently folded C-terminal domain of influenza polymerase subunit PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Boudet, J, Tarendeau, F, Guilligay, D, Mas, P, Bougault, C.M, Cusack, S, Simorre, J.-P, Hart, D.J. | | Deposit date: | 2006-04-07 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1O15
 
 | |
2MHK
 
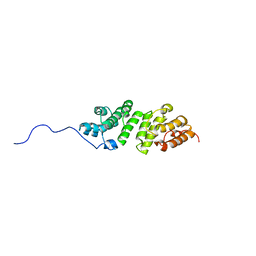 | | E. coli LpoA N-terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Jean, N.L, Bougault, C, Lodge, A, Derouaux, A, Callens, G, Egan, A, Lewis, R.J, Vollmer, W, Simorre, J. | | Deposit date: | 2013-11-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Structure, 22, 2014
|
|
4A52
 
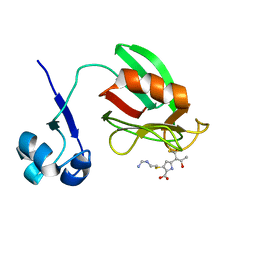 | | NMR structure of the imipenem-acylated L,D-transpeptidase from Bacillus subtilis | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, PUTATIVE L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J, Bougault, C, Arthur, M, Hugonnet, J, Veckerle, C, Pessey, O. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-30 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
