4JIW
 
 | | c1882 PAAR-repeat protein from Escherichia coli in complex with a VgrG-like beta-helix that is based on a fragment of T4 gp5 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative uncharacterized protein, ... | | Authors: | Buth, S.A, Leiman, P.G, Shneider, M.M. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | PAAR-repeat proteins sharpen and diversify the type VI secretion system spike.
Nature, 500, 2013
|
|
4JIV
 
 | | VCA0105 PAAR-repeat protein from Vibrio cholerae in complex with a VgrG-like beta-helix that is based on a fragment of T4 gp5 | | Descriptor: | 9-OCTADECENOIC ACID, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Buth, S.A, Leiman, P.G, Shneider, M.M. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | PAAR-repeat proteins sharpen and diversify the type VI secretion system spike.
Nature, 500, 2013
|
|
6P20
 
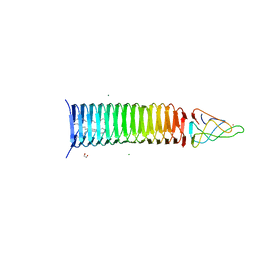 | | Bacteriophage phiKZ gp163.1 PAAR repeat protein in complex with a T4 gp5 beta-helix fragment modified to mimic the phiKZ central spike gp164 | | Descriptor: | 1,2-ETHANEDIOL, 9-OCTADECENOIC ACID, Baseplate central spike complex protein gp5,PHIKZ164, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2019-05-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Bacteriophage phiKZ gp163.1 PAAR repeat protein in complex with a T4 gp5 beta-helix fragment modified to mimic the phiKZ central spike gp164
To Be Published
|
|
6P1Z
 
 | |
6P22
 
 | | Photorhabdus Virulence Cassette (PVC) PAAR repeat protein Pvc10 in complex with a T4 gp5 beta-helix fragment modified to mimic Pvc8, the central spike protein of PVC | | Descriptor: | 9-OCTADECENOIC ACID, CHIMERA OF CENTRAL SPIKE PROTEINS GP5 FROM PHAGE T4 AND PVC8 FROM PVC, MAGNESIUM ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2019-05-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Photorhabdus Virulence Cassette (PVC) PAAR repeat protein Pvc10 in complex with a T4 gp5 beta-helix fragment modified to mimic Pvc8, the central spike protein of PVC
To Be Published
|
|
6P2A
 
 | |
5IW9
 
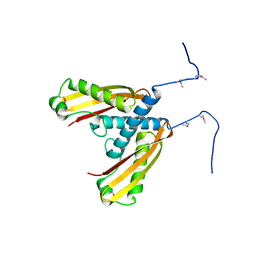 | |
3BKV
 
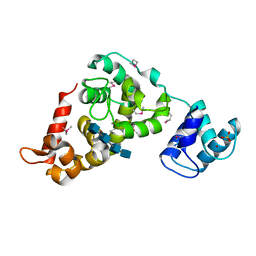 | | X-ray structure of the bacteriophage phiKZ lytic transglycosylase, gp144, in complex with chitotetraose, (NAG)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Fokine, A, Miroshnikov, K.A, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2007-12-07 | | Release date: | 2008-01-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the bacteriophage phi KZ lytic transglycosylase gp144.
J.Biol.Chem., 283, 2008
|
|
3BKH
 
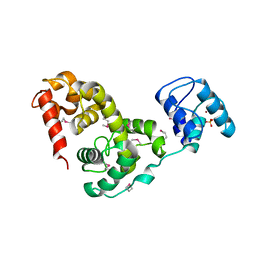 | | Crystal structure of the bacteriophage phiKZ lytic transglycosylase, gp144 | | Descriptor: | NICKEL (II) ION, SULFATE ION, lytic transglycosylase | | Authors: | Fokine, A, Miroshnikov, K.A, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the bacteriophage phi KZ lytic transglycosylase gp144.
J.Biol.Chem., 283, 2008
|
|
4MTK
 
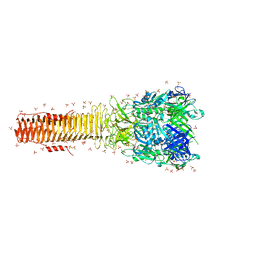 | |
4MTM
 
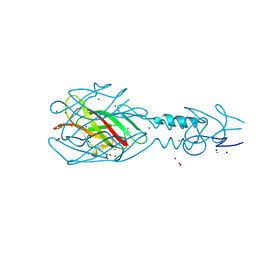 | | Crystal structure of the tail fiber gp53 from Acinetobacter baumannii bacteriophage AP22 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GLYCEROL, ... | | Authors: | Sycheva, L.V, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2013-09-19 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.368 Å) | | Cite: | Crystal Structure of the putative tail fiber protein gp53 from the Acinetobacter baumannii bacteriophage AP22
Biorxiv, 2019
|
|
4Y9V
 
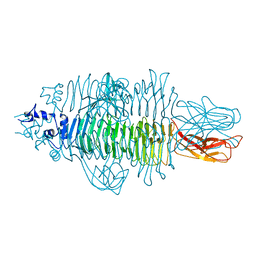 | | Gp54 tailspike of Acinetobacter baumannii bacteriophage AP22 in complex with A. baumannii capsular saccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2,4-dideoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-fucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-mannopyranuronic acid, CHLORIDE ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2015-02-17 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of Acinetobacter baumannii bacteriophage AP22 polysaccharide degrading lyase in complex with A. baumannii capsular saccharide at 0.9 A resolution
TO BE PUBLISHED
|
|
6U5J
 
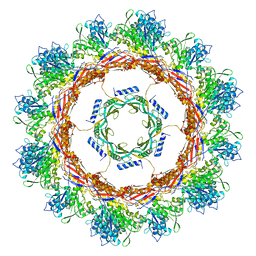 | | CryoEM Structure of Pyocin R2 - postcontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
5CES
 
 | |
4GBF
 
 | |
6U5B
 
 | | CryoEM Structure of Pyocin R2 - precontracted - baseplate | | Descriptor: | Glue PA0627, Ripcord PA0626, Sheath Initiator PA0617, ... | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6U5H
 
 | | CryoEM Structure of Pyocin R2 - precontracted - hub | | Descriptor: | Probable bacteriophage protein Pyocin R2 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6U5K
 
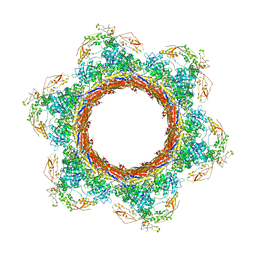 | | CryoEM Structure of Pyocin R2 - postcontracted - baseplate | | Descriptor: | Glue PA0627, Sheath Initiator PA0617, Sheath PA0622, ... | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6U5F
 
 | | CryoEM Structure of Pyocin R2 - precontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622, Tube PA0623 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
2FKK
 
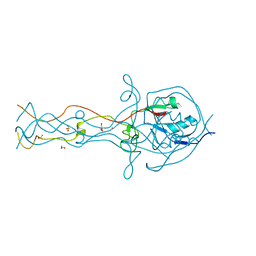 | | Crystal structure of the C-terminal domain of the bacteriophage T4 gene product 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2006-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Evolution of bacteriophage tails: structure of t4 gene product 10
J.Mol.Biol., 358, 2006
|
|
2FL9
 
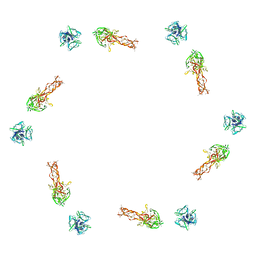 | |
3H2T
 
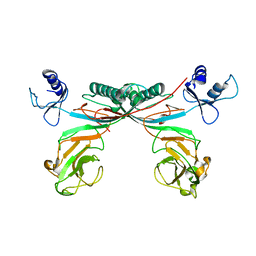 | | Crystal structure of gene product 6, baseplate protein of bacteriophage T4 | | Descriptor: | Baseplate structural protein Gp6 | | Authors: | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2009-04-14 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of gene product 6 of bacteriophage t4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
3H3W
 
 | | Fitting of the gp6 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 dome-shaped baseplate | | Descriptor: | Baseplate structural protein Gp6 | | Authors: | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
3H3Y
 
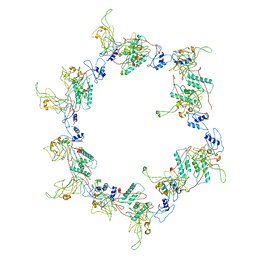 | | Fitting of the gp6 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 star-shaped baseplate | | Descriptor: | Baseplate structural protein Gp6 | | Authors: | Aksyuk, A.A, Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate.
Structure, 17, 2009
|
|
6ET6
 
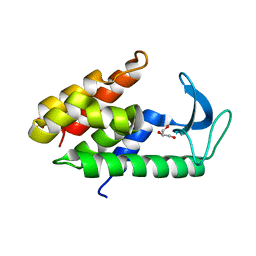 | | Crystal structure of muramidase from Acinetobacter baumannii AB 5075UW prophage | | Descriptor: | GLYCEROL, Lysozyme, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Sykilinda, N.N, Shneider, M.M, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2017-10-25 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of anAcinetobacterBroad-Range Prophage Endolysin Reveals a C-Terminal alpha-Helix with the Proposed Role in Activity against Live Bacterial Cells.
Viruses, 10, 2018
|
|
