7D6I
 
 | | A neutralizing MAb targeting receptor-binding-domain of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of GH12-Fab, Light chain of GH12-Fab, ... | | Authors: | Shi, R, Qi, J.X, Gao, G.F, Yan, J.H. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | A neutralizing MAb targeting receptor-binding-domain of SARS-CoV-2
To Be Published
|
|
3CES
 
 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | Descriptor: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
3G05
 
 | | Crystal structure of N-terminal domain (2-550) of E.coli MnmG | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
3LVL
 
 | | Crystal Structure of E.coli IscS-IscU complex | | Descriptor: | Cysteine desulfurase, NifU-like protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LA0
 
 | | Crystal Structure of UreE from Helicobacter pylori (metal of unknown identity bound) | | Descriptor: | UNKNOWN ATOM OR ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3L9Z
 
 | | Crystal Structure of UreE from Helicobacter pylori (apo form) | | Descriptor: | Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3LVJ
 
 | | Crystal Structure of E.coli IscS-TusA complex (form 1) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVK
 
 | | Crystal Structure of E.coli IscS-TusA complex (form 2) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LVM
 
 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3NY0
 
 | | Crystal Structure of UreE from Helicobacter pylori (Ni2+ bound form) | | Descriptor: | NICKEL (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3NXZ
 
 | | Crystal Structure of UreE from Helicobacter pylori (Cu2+ bound form) | | Descriptor: | COPPER (II) ION, Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3CQJ
 
 | |
3CQK
 
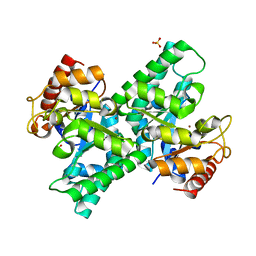 | | Crystal Structure of L-xylulose-5-phosphate 3-epimerase UlaE (form B) complex with Zn2+ and sulfate | | Descriptor: | L-ribulose-5-phosphate 3-epimerase ulaE, SULFATE ION, ZINC ION | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of L-xylulose-5-Phosphate 3-epimerase (UlaE) from the anaerobic L-ascorbate utilization pathway of Escherichia coli: identification of a novel phosphate binding motif within a TIM barrel fold.
J.Bacteriol., 190, 2008
|
|
3CQI
 
 | |
3CQH
 
 | |
3G2Q
 
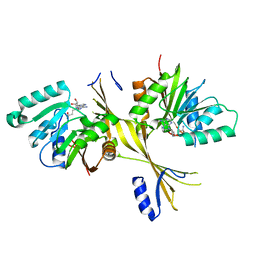 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with sinefungin | | Descriptor: | PCZA361.24, SINEFUNGIN | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
3G2P
 
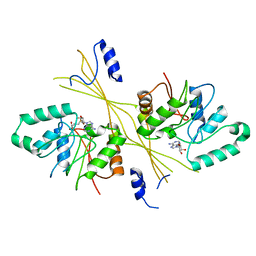 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-homocysteine (SAH) | | Descriptor: | PCZA361.24, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
3G2M
 
 | |
7C01
 
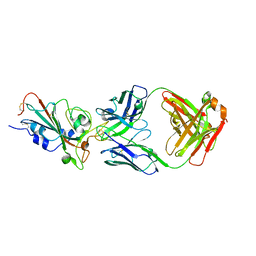 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
4HE0
 
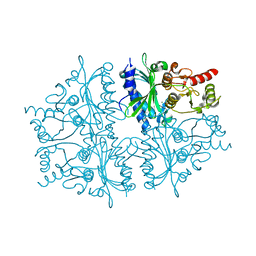 | | Crystal structure of human muscle fructose-1,6-bisphosphatase | | Descriptor: | CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, MAGNESIUM ION, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3G2O
 
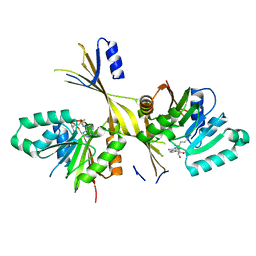 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-methionine (SAM) | | Descriptor: | PCZA361.24, S-ADENOSYLMETHIONINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
4EEC
 
 | |
4HE2
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
4HE1
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with fructose-6-phosphate and phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
3PNL
 
 | | Crystal Structure of E.coli Dha kinase DhaK-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
