5NWM
 
 | |
5NWX
 
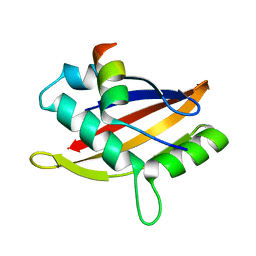 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
8P7G
 
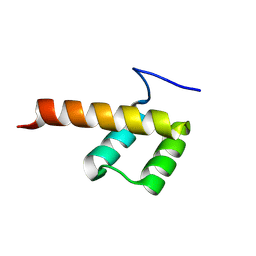 | |
2MYL
 
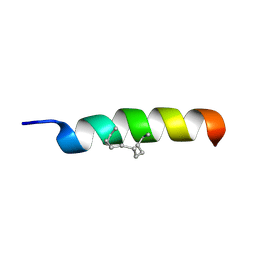 | |
2MYM
 
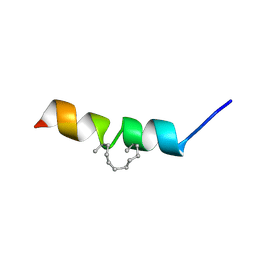 | |
2M97
 
 | |
3TEO
 
 | | APO Form of carbon disulfide hydrolase (selenomethionine form) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, Carbon disulfide hydrolase | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
3TEN
 
 | | Holo form of carbon disulfide hydrolase | | Descriptor: | CS2 hydrolase, ZINC ION | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
2MLG
 
 | | Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx | | Descriptor: | Sulfolobus transcription factor 76 aminoacid protein, Stf76 | | Authors: | Farina, B, Russo, L, Fattorusso, R. | | Deposit date: | 2014-02-27 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx: a novel peculiar member of the winged helix-turn-helix transcription factor family.
Nucleic Acids Res., 42, 2014
|
|
2LQY
 
 | | Structure and orientation of the gH625-644 membrane interacting region of herpes simplex virus type 1 in a membrane mimetic system. | | Descriptor: | Envelope glycoprotein H | | Authors: | Isernia, C, Galdiero, S, Russo, L, Falanga, A, Cantisani, M, Vitiello, M, Fattorusso, R, Malgieri, G, Galdiero, M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Orientation of the gH625-644 Membrane Interacting Region of Herpes Simplex Virus Type 1 in a Membrane Mimetic System.
Biochemistry, 51, 2012
|
|
1NBA
 
 | | CRYSTAL STRUCTURE ANALYSIS, REFINEMENT AND ENZYMATIC REACTION MECHANISM OF N-CARBAMOYLSARCOSINE AMIDOHYDROLASE FROM ARTHROBACTER SP. AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | N-CARBAMOYLSARCOSINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Romao, M.J, Turk, D, Gomis-Ruth, F.-Z, Huber, R, Schumacher, G, Mollering, H, Russmann, L. | | Deposit date: | 1992-05-18 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
2N5Z
 
 | |
5OLF
 
 | |
2L1O
 
 | | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2His2 zinc finger induces structural rearrangements of typical DNA base determinant positions | | Descriptor: | CADMIUM ION, Transcriptional regulator SUPERMAN | | Authors: | Malgieri, G, Zaccaro, L, Leone, M, Bucci, E, Esposito, S, Baglivo, I, Del Gatto, A, Scandurra, R, Pedone, P.V, Fattorusso, R, Isernia, C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-08 | | Last modified: | 2011-12-21 | | Method: | SOLUTION NMR | | Cite: | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys(2) His(2) zinc finger induces structural rearrangements of typical DNA base determinant positions.
Biopolymers, 95, 2011
|
|
2JSP
 
 | |
1CHM
 
 | | ENZYMATIC MECHANISM OF CREATINE AMIDINOHYDROLASE AS DEDUCED FROM CRYSTAL STRUCTURES | | Descriptor: | CARBAMOYL SARCOSINE, CREATINE AMIDINOHYDROLASE | | Authors: | Hoeffken, H.W, Knof, S.H, Bartlett, P.A, Huber, R, Moellering, H, Schumacher, G. | | Deposit date: | 1993-07-19 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures.
J.Mol.Biol., 214, 1990
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
