2QK7
 
 | | A covalent S-F heterodimer of staphylococcal gamma-hemolysin | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Roblin, P, Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2007-07-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of beta-barrel pore-forming toxins.
Proteins, 71, 2008
|
|
3J4J
 
 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | Deposit date: | 2013-08-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4WKR
 
 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | Descriptor: | 7SK GGHP4 (300-332), La-related protein 7 | | Authors: | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | Deposit date: | 2014-10-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
1T5R
 
 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN S COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | LukS-PV | | Authors: | Guillet, V, Roblin, P, Keller, D, Prevost, G, Mourey, L. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of leucotoxin S component: new insight into the Staphylococcal beta-barrel pore-forming toxins.
J.Biol.Chem., 279, 2004
|
|
2N5D
 
 | | NMR structure of PKS domains | | Descriptor: | fusion protein of two PKS domains | | Authors: | Dorival, J, Annaval, T, Risser, F, Collin, S, Roblin, P, Jacob, C, Gruez, A, Chagot, B, Weissman, K.J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
6GYV
 
 | | Lariat-capping ribozyme (circular permutation form) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lariat-capping ribozyme, MAGNESIUM ION, ... | | Authors: | Masquida, B, Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E. | | Deposit date: | 2018-07-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.50003624 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
5OLL
 
 | | Crystal structure of gurmarin, a sweet taste suppressing polypeptide | | Descriptor: | Gurmarin, NICKEL (II) ION | | Authors: | Sigoillot, M, Neiers, F, Legrand, P, Roblin, P, Briand, L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Gurmarin, a Sweet Taste-Suppressing Protein: Identification of the Amino Acid Residues Essential for Inhibition.
Chem. Senses, 43, 2018
|
|
4P9R
 
 | | Speciation of a group I intron into a lariat capping ribozyme (Heavy atom derivative) | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (189-MER) | | Authors: | Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E, Masquida, B. | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P8Z
 
 | | Speciation of a group I intron into a lariat capping ribozyme | | Descriptor: | Didymium iridis partial IGS, 18S rRNA gene, I-DirI gene and partial ITS1 | | Authors: | Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E, Masquida, B. | | Deposit date: | 2014-04-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4UDJ
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3SJH
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP-Latrunculin A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-06-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
5FT9
 
 | | Arabidopsis thaliana nuclear protein-only RNase P 2 (PRORP2) | | Descriptor: | PROTEINACEOUS RNASE P 2, ZINC ION | | Authors: | Fernandez-Millan, P, Pinker, F, Schelcher, C, Gobert, A, Giege, P, Sauter, C. | | Deposit date: | 2016-01-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of Arabidopsis Nuclear Rnase P Alone and with tRNA Reveal Plasticities
To be Published
|
|
4UDK
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.76 Angstrom from unknown human gut bacteria (Uhgb_MP) in complex with N-acetyl-D-glucosamine, beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDI
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.85 Angstrom from unknown human gut bacteria (Uhgb_MP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3N5C
 
 | | Crystal Structure of Arf6DELTA13 complexed with GDP | | Descriptor: | ADP-ribosylation factor 6, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Aizel, K, Biou, V, Cherfils, J. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6.
J.Mol.Biol., 402, 2010
|
|
5LFC
 
 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M | | Descriptor: | CALCIUM ION, DsrV, GLYCEROL | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S. | | Deposit date: | 2016-07-01 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 7, 2017
|
|
5DOL
 
 | | Crystal structure of YabA amino-terminal domain from Bacillus subtilis | | Descriptor: | Initiation-control protein YabA | | Authors: | Cherrier, M.V, Bazin, A, Jameson, K.H, Wilkinson, A.J, Noirot-Gros, M.F, Terradot, L. | | Deposit date: | 2015-09-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners.
Nucleic Acids Res., 44, 2016
|
|
4UDG
 
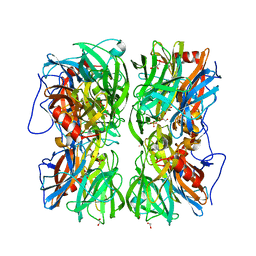 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with N-acetylglucosamine and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3U8X
 
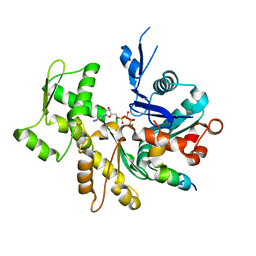 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3U9Z
 
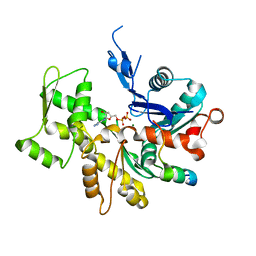 | | Crystal structure between actin and a protein construct containing the first beta-thymosin domain of drosophila ciboulot (residues 2-58) with the three mutations N26D/Q27K/D28S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3U9D
 
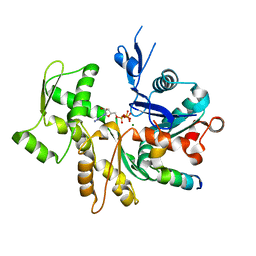 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-24) of drosophila Ciboulot and the C-terminal domain (residues 13-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly.
Embo J., 31, 2012
|
|
4RQI
 
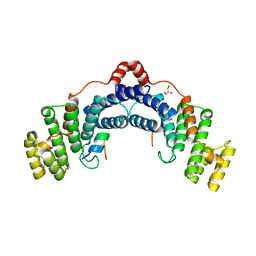 | | Structure of TRF2/RAP1 secondary interaction binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, Telomeric repeat-binding factor 2, ... | | Authors: | Miron, S, Guimaraes, B, Gaullier, G, Giraud-Panis, M.-J, Gilson, E, Le Du, M.-H. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4405 Å) | | Cite: | A higher-order entity formed by the flexible assembly of RAP1 with TRF2.
Nucleic Acids Res., 44, 2016
|
|
5NGY
 
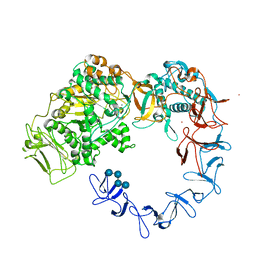 | | Crystal structure of Leuconostoc citreum NRRL B-1299 dextransucrase DSR-M | | Descriptor: | CALCIUM ION, DSR-M glucansucrase inactive mutant E715Q, PRASEODYMIUM ION, ... | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S, Vuillemin, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 2017
|
|
5O8L
 
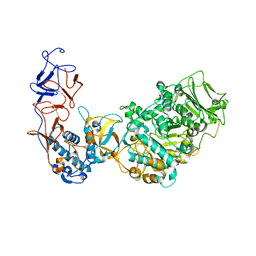 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with sucrose | | Descriptor: | Alternansucrase, CALCIUM ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S. | | Deposit date: | 2017-06-13 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 7, 2017
|
|
6G7Z
 
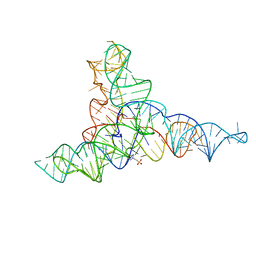 | |
