6LYE
 
 | | Crystal Structure of mimivirus UNG Y322F in complex with UGI | | Descriptor: | Probable uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
J.Struct.Biol., 211, 2020
|
|
6LYD
 
 | | Crystal Structure of mimivirus UNG Y322L in complex with UGI | | Descriptor: | Probable uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
J.Struct.Biol., 211, 2020
|
|
6M37
 
 | |
6M36
 
 | |
1DIN
 
 | |
7WA4
 
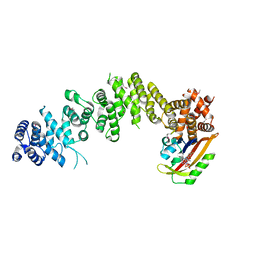 | | Crystal structure of GIGANTEA in complex with LKP2 | | Descriptor: | Adagio protein 2, FLAVIN MONONUCLEOTIDE, Protein GIGANTEA | | Authors: | Pathak, D, Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the regulation of blue-light receptors by GIGANTEA.
Cell Rep, 39, 2022
|
|
8ACT
 
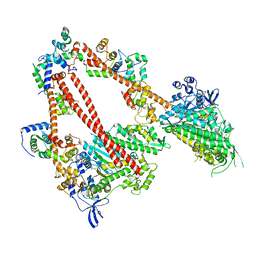 | | structure of the human beta-cardiac myosin folded-back off state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | Authors: | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | Deposit date: | 2022-07-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
5X55
 
 | |
8WWZ
 
 | |
7WSJ
 
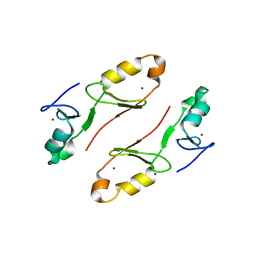 | | Crystal structure of the tandem B-box domain of Arabidopsis thaliana CONSTANS | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS | | Authors: | Dahal, P, Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2022-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a tandem B-box domain from Arabidopsis CONSTANS.
Biochem.Biophys.Res.Commun., 599, 2022
|
|
6JHK
 
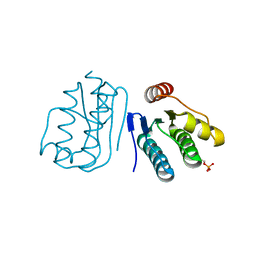 | |
6JHE
 
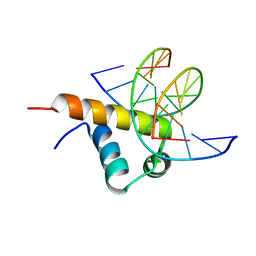 | | Crystal Structure of Bacillus subtilis SigW domain 4 in complexed with -35 element DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*AP*AP*CP*CP*TP*TP*T)-3'), ECF RNA polymerase sigma factor SigW | | Authors: | Kwon, E, Devkota, S.R, Pathak, D, Dahal, P, Kim, D.Y. | | Deposit date: | 2019-02-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural analysis of the recognition of the -35 promoter element by SigW from Bacillus subtilis.
Plos One, 14, 2019
|
|
5TXK
 
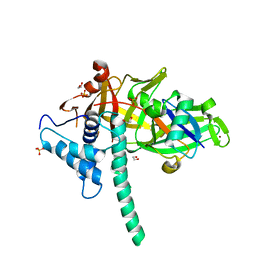 | | CRYSTAL STRUCTURE OF USP35 C450S IN COMPLEX WITH UBIQUITIN | | Descriptor: | 1,2-ETHANEDIOL, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Bader, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expansion of DUB functionality generated by alternative isoforms - USP35, a case study.
J. Cell. Sci., 131, 2018
|
|
