3BF7
 
 | |
3BF8
 
 | |
3BD3
 
 | |
3BD5
 
 | |
3BD4
 
 | |
6LUN
 
 | | NN2101 Antibody Fab fragment | | Descriptor: | NN2101 | | Authors: | Kim, H.N, Seo, M.D, Park, S.K. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development and characterization of a fully human antibody targeting SCF/c-kit signaling.
Int.J.Biol.Macromol., 159, 2020
|
|
5GLT
 
 | | Tl-gal with LNT | | Descriptor: | Galectin, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jang, S.B, Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
5GM0
 
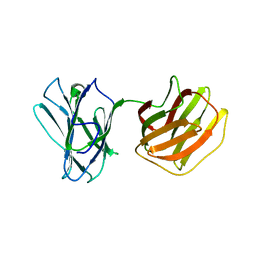 | | Tl-gal with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
5GLV
 
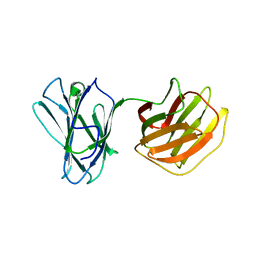 | | Tl-gal | | Descriptor: | galectin | | Authors: | Jang, S.B, Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
5GLW
 
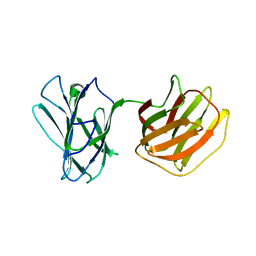 | | Tl-gal with LacNAc | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin | | Authors: | Jang, S.B, Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
5GLU
 
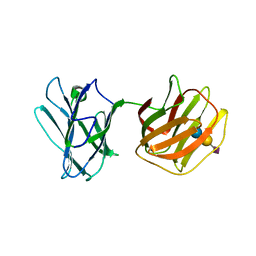 | | Tl-gal with SiaLac | | Descriptor: | GALECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jang, S.B, Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
5GLZ
 
 | | Tl-gal with Glucose | | Descriptor: | beta-D-glucopyranose, galectin | | Authors: | Jang, S.B, Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
5Z9G
 
 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5Z9H
 
 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
