7YQ2
 
 | | Crystal structure of photosystem II expressing psbA2 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
7YQ7
 
 | | Crystal structure of photosystem II expressing psbA3 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
1V93
 
 | | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5,10-Methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakajima, Y, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of 5,10-Methylenetetrahydrofolate reductase from Thermus thermophilus HB8
To be Published
|
|
1IS2
 
 | |
1X2E
 
 | | The crystal structure of prolyl aminopeptidase complexed with Ala-TBODA | | Descriptor: | (2S)-2-AMINO-1-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)PROPAN-1-ONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Ito, K, Sakata, M, Xu, Y, Matsubara, F, Hatakeyama, S, Yoshimoto, T. | | Deposit date: | 2005-04-22 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual extra space at the active site and high activity for acetylated hydroxyproline of prolyl aminopeptidase from Serratia marcescens
J.Bacteriol., 188, 2006
|
|
1X2B
 
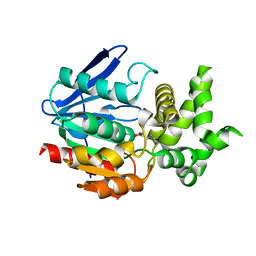 | | The crystal structure of prolyl aminopeptidase complexed with Sar-TBODA | | Descriptor: | 1-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-2-(METHYLAMINO)ETHANONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Ito, K, Sakata, M, Xu, Y, Matsubara, F, Hatakeyama, S, Yoshimoto, T. | | Deposit date: | 2005-04-22 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual extra space at the active site and high activity for acetylated hydroxyproline of prolyl aminopeptidase from Serratia marcescens
J.Bacteriol., 188, 2006
|
|
7VKF
 
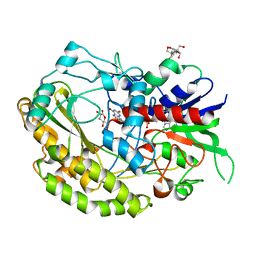 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y, Nishiya, Y, Ito, K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VKD
 
 | |
7VZS
 
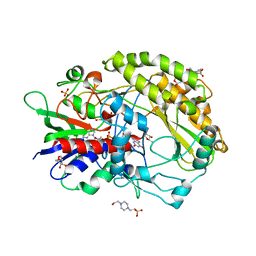 | | FAD-dpendent Glucose Dehydrogenase complexed with an inhibitor at pH7.56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-glucal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VZP
 
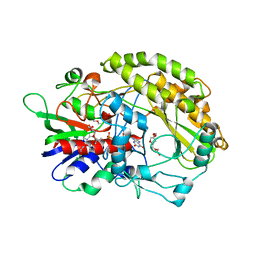 | | FAD-dpendent Glucose Dehydrogenase from Aspergillus oryzae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase, PENTAETHYLENE GLYCOL | | Authors: | Nakajima, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
2ECF
 
 | |
5ZZN
 
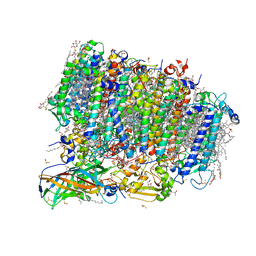 | | Crystal structure of photosystem II from an SQDG-deficient mutant of Thermosynechococcus elongatus | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Nakajima, Y, Umena, Y, Nagao, R, Endo, K, Kobayashi, K, Akita, F, Suga, M, Wada, H, Noguchi, T, Shen, J.R. | | Deposit date: | 2018-06-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thylakoid membrane lipid sulfoquinovosyl-diacylglycerol (SQDG) is required for full functioning of photosystem II inThermosynechococcus elongatus.
J. Biol. Chem., 293, 2018
|
|
2DCM
 
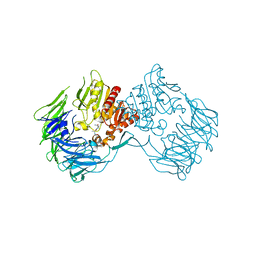 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2D5L
 
 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2DQ6
 
 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
3A6J
 
 | | E122Q mutant creatininase complexed with creatine | | Descriptor: | Creatinine amidohydrolase, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6D
 
 | | Creatininase complexed with 1-methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6L
 
 | | E122Q mutant creatininase, Zn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6H
 
 | | W154A mutant creatininase | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6F
 
 | | W174F mutant creatininase, Type II | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6G
 
 | | W154F mutant creatininase | | Descriptor: | Creatinine amidohydrolase, MANGANESE (II) ION, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
2ZXG
 
 | | Aminopeptidase N complexed with the aminophosphinic inhibitor of PL250, a transition state analogue | | Descriptor: | Aminopeptidase N, GLYCEROL, N-{(2S)-3-[(1R)-1-aminoethyl](hydroxy)phosphoryl-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli complexed with the transition-state analogue aminophosphinic inhibitor PL250
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4UB6
 
 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
