3RAP
 
 | |
2J59
 
 | | Crystal structure of the ARF1:ARHGAP21-ArfBD complex | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ADP-RIBOSYLATION FACTOR 1, ... | | Authors: | Menetrey, J, Perderiset, M, Cicolari, J, Dubois, T, El Khatib, N, El Khadali, F, Franco, M, Chavrier, P, Houdusse, A. | | Deposit date: | 2006-09-13 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Arf1-Mediated Recruitment of Arhgap21 to Golgi Membranes.
Embo J., 26, 2007
|
|
1GZE
 
 | | Structure of the Clostridium botulinum C3 exoenzyme (L177C mutant) | | Descriptor: | MERCURY (II) ION, MONO-ADP-RIBOSYLTRANSFERASE C3 | | Authors: | Menetrey, J, Flatau, G, Stura, E.A, Charbonnier, J.B, Gas, F, Teulon, J.M, Le Du, M.H, Boquet, P, Menez, A. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nad Binding Induces Conformational Changes in Rho Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme
J.Biol.Chem., 277, 2002
|
|
1GZF
 
 | | Structure of the Clostridium botulinum C3 exoenzyme (wild-type) in complex with NAD | | Descriptor: | 3-(AMINOCARBONYL)-1-[(3R,4S,5R)-3,4-DIHYDROXY-5-METHYLTETRAHYDRO-2-FURANYL]PYRIDINIUM, ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Menetrey, J, Flatau, G, Stura, E.A, Charbonnier, J.B, Gas, F, Teulon, J.M, Le Du, M.H, Boquet, P, Menez, A. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nad Binding Induces Conformational Changes in Rho Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme
J.Biol.Chem., 277, 2002
|
|
8OUK
 
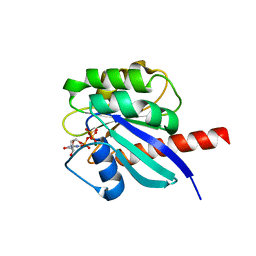 | | Arf GTPase from the asgard Hodarchaea : HodArfR1 bound to GTP | | Descriptor: | Arf GTPase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Menetrey, J, Jackson, C, Dacks, J, Elias, M, Vargova, R. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arf Family GTPases are present in Asgard archaea
To Be Published
|
|
8OUL
 
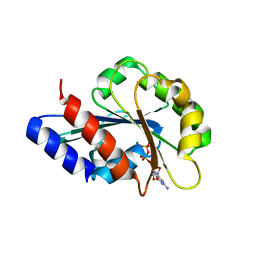 | |
8OUM
 
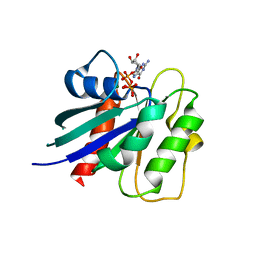 | | Arf GTPase from the asgard Gerdarchaea : GerdArfR1 bound to GTP | | Descriptor: | GTP-binding protein, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Menetrey, J, Jackson, C, Dacks, J.B, Elias, M, Vargova, R. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Arf Family GTPases are present in Asgard archaea
To Be Published
|
|
8OUN
 
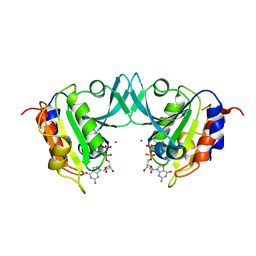 | | Arf GTPase from the asgard Gerdarchaea : GerdArfR1 bound to GDP | | Descriptor: | GTP-binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Menetrey, J, Jackson, C, Dacks, J.B, Elias, M, Vargova, R. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Arf Family GTPases are present in Asgard archaea
To Be Published
|
|
4ANJ
 
 | | MYOSIN VI (MDinsert2-GFP fusion) PRE-POWERSTROKE STATE (MG.ADP.AlF4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Menetrey, J, Isabet, T, Ropars, V, Mukherjea, M, Pylypenko, O, Liu, X, Perez, J, Vachette, P, Sweeney, H.L, Houdusse, A.M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin Vi.
Mol.Cell, 48, 2012
|
|
1E0S
 
 | | small G protein Arf6-GDP | | Descriptor: | ADP-ribosylation factor 6, AMMONIUM ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Menetrey, J, Cherfils, J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of Arf6-Gdp Suggests a Basis for Guanine Nucleotide Exchange Factors Specificity
Nat.Struct.Biol., 7, 2000
|
|
2VAS
 
 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
2VB6
 
 | | Myosin VI (MD-insert2-CaM, Delta Insert1) Post-rigor state (crystal form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
7AI4
 
 | |
7AIE
 
 | |
2V26
 
 | | Myosin VI (MD) pre-powerstroke state (Mg.ADP.VO4) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Mukherjea, M, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-06-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structural Basis for the Large Powerstroke of Myosin Vi.
Cell(Cambridge,Mass.), 131, 2007
|
|
2BKI
 
 | | Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure | | Descriptor: | CALCIUM ION, CALMODULIN, SULFATE ION, ... | | Authors: | Menetrey, J, Bahloul, A, Yengo, C, Wells, A, Morris, C, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of the Myosin Vi Motor Reveals the Mechanism of Directionality Reversal
Nature, 435, 2005
|
|
2BKH
 
 | | Myosin VI nucleotide-free (MDInsert2) crystal structure | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Menetrey, J, Bahloul, A, Yengo, C, Wells, A, Morris, C, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the Myosin Vi Motor Reveals the Mechanism of Directionality Reversal
Nature, 435, 2005
|
|
2C8F
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (NAD-bound state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8A
 
 | | Structure of the wild-type C3bot1 Exoenzyme (Nicotinamide-bound state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8C
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C89
 
 | | Structure of the wild-type C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8D
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
5OJF
 
 | | Crystal Structure of KLC2-TPR domain (fragment [A1-B6] | | Descriptor: | Kinesin light chain 2 | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Andreani, J, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
