5AQ6
 
 | | Structure of E. coli ZinT at 1.79 Angstrom | | Descriptor: | ACETIC ACID, METAL-BINDING PROTEIN ZINT, ZINC ION | | Authors: | Santo, P.E, Colaco, H.G, Matias, P, Vicente, J.B, Bandeiras, T.M. | | Deposit date: | 2015-09-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Roles of Escherichia Coli Zint in Cobalt, Mercury and Cadmium Resistance and Structural Insights Into the Metal Binding Mechanism
Metallomics, 8, 2016
|
|
8C4L
 
 | |
5J2V
 
 | | Crystal Structure of Hsp90-alpha Apo N-domain | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J64
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(2,4-dihydroxyphenyl)-4-(2-fluorophenyl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J20
 
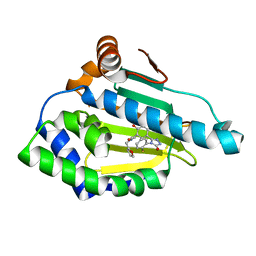 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J6L
 
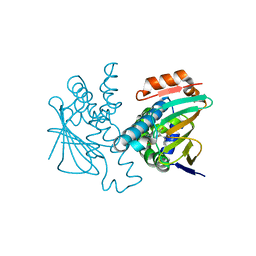 | | Crystal Structure of Hsp90-alpha N-domain in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J86
 
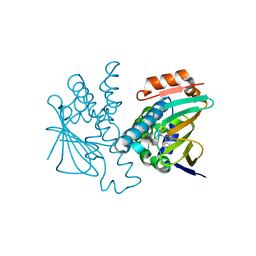 | | Crystal Structure of Hsp90-alpha N-domain in complex with 2,4-Dihydroxy-N-methyl-5-(5-oxo-4-o-tolyl-4,5-dihydro-1H-[1,2,4]triazol-3-yl)-N-thiophen-2-ylmethyl-benzamide | | Descriptor: | 2,4-dihydroxy-N-methyl-5-[4-(2-methylphenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(thiophen-2-yl)methyl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J2X
 
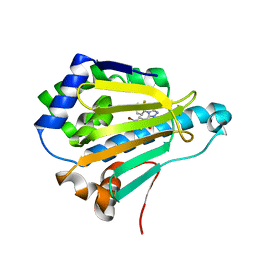 | | Crystal Structure of Hsp90-alpha N-domain in complex with 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J6M
 
 | | Crystal Structure of Hsp90-alpha N-domain L107 mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J8M
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J8U
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J27
 
 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J6N
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J82
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-isopropyl-N-methyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-(propan-2-yl)benzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J9X
 
 | | HSP90 in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-11 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J80
 
 | |
1E5D
 
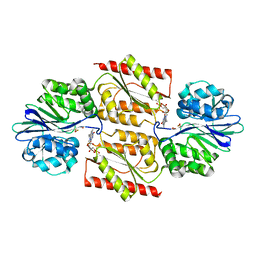 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
7OYI
 
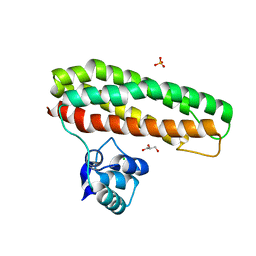 | | Escherichia coli YtfE_E159L(MN) | | Descriptor: | CHLORIDE ION, GLYCEROL, Iron-sulfur cluster repair protein YtfE, ... | | Authors: | Silva, L.S.O, Matias, P, Romao, C.V, Saraiva, L.M. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Repair of Iron Center Proteins-A Different Class of Hemerythrin-like Proteins.
Molecules, 27, 2022
|
|
6GEH
 
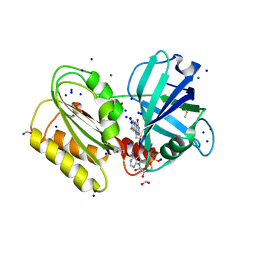 | | Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility. | | Descriptor: | DIMETHYL SULFOXIDE, FAD-binding 9, siderophore-interacting domain protein, ... | | Authors: | Trindade, I.B, Silva, J.P.M, Matias, P, Moe, E. | | Deposit date: | 2018-04-26 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and reactivity of a siderophore-interacting protein from the marine bacteriumShewanellareveals unanticipated functional versatility.
J. Biol. Chem., 294, 2019
|
|
6HR0
 
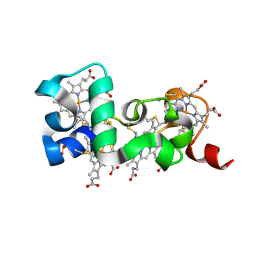 | |
6H7X
 
 | | First X-ray structure of full-length human RuvB-Like 2. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Silva, S, Brito, J.A, Matias, P, Bandeiras, T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | X-ray structure of full-length human RuvB-Like 2 - mechanistic insights into coupling between ATP binding and mechanical action.
Sci Rep, 8, 2018
|
|
1DFX
 
 | | DESULFOFERRODOXIN FROM DESULFOVIBRIO DESULFURICANS, ATCC 27774 | | Descriptor: | CALCIUM ION, DESULFOFERRODOXIN, FE (III) ION | | Authors: | Coelho, A.V, Matias, P.M, Carrondo, M.A. | | Deposit date: | 1997-09-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Desulfoferrodoxin Structure Determined by MAD Phasing and Refinement to 1.9 Angstroms Resolution Reveals a Unique Combination of a Tetrahedral Fes4 Centre with a Square Pyramidal Fesn4 Centre
J.Biol.Inorg.Chem., 2, 1997
|
|
4C4U
 
 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus E12Q mutant in the reduced form | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | Deposit date: | 2013-09-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Understanding the Role of Key Residues in the Superoxide Reductase Molecular Mechanism, Exploring Archaeoglobus Fulgidus Sor Structure
To be Published
|
|
8RE3
 
 | | Crystal Structure determination of Dye-decolorizing Peroxidase (DyP) mutant M190G from Deinoccoccus radiodurans | | Descriptor: | CALCIUM ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Salgueiro, B.A, Frade, K, Frazao, C, Matias, P, Moe, E. | | Deposit date: | 2023-12-10 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical, Biophysical, and Structural Analysis of an Unusual DyP from the Extremophile Deinococcus radiodurans.
Molecules, 29, 2024
|
|
