1FMF
 
 | | REFINED SOLUTION STRUCTURE OF THE (13C,15N-LABELED) B12-BINDING SUBUNIT OF GLUTAMATE MUTASE FROM CLOSTRIDIUM TETANOMORPHUM | | Descriptor: | METHYLASPARTATE MUTASE S CHAIN | | Authors: | Hoffmann, B, Konrat, R, Tollinger, M, Huhta, M, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 2000-08-17 | | Release date: | 2002-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A protein pre-organized to trap the nucleotide moiety of coenzyme B(12): refined solution structure of the B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum.
Chembiochem, 2, 2001
|
|
1LQ9
 
 | | Crystal Structure of a Monooxygenase from the Gene ActVA-Orf6 of Streptomyces coelicolor Strain A3(2) | | Descriptor: | ACTVA-ORF6 MONOOXYGENASE, TETRAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
EMBO J., 22, 2003
|
|
1ID8
 
 | | NMR STRUCTURE OF GLUTAMATE MUTASE (B12-BINDING SUBUNIT) COMPLEXED WITH THE VITAMIN B12 NUCLEOTIDE | | Descriptor: | 2-HYDROXY-PROPYL-AMMONIUM, METHYLASPARTATE MUTASE S CHAIN, PHOSPHORIC ACID MONO-[5-(5,6-DIMETHYL-BENZOIMIDAZOL-1-YL)-4-HYDROXY-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER | | Authors: | Tollinger, M, Eichmuller, C, Konrat, R, Huhta, M.S, Marsh, E.N.G, Krautler, B. | | Deposit date: | 2001-04-04 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum traps the nucleotide moiety of coenzyme B(12).
J.Mol.Biol., 309, 2001
|
|
1BE1
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Tollinger, M, Konrat, R, Hilbert, B.H, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum
Structure, 6, 1998
|
|
4TW3
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4G4L
 
 | | Crystal structure of the de novo designed peptide alpha4tbA6 | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
4G3B
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3d | | Descriptor: | ACETYL GROUP, alpha4F3d | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-13 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
4G4M
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3(6-13) | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, alpha4F3(6-13) | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | Descriptor: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWE
 
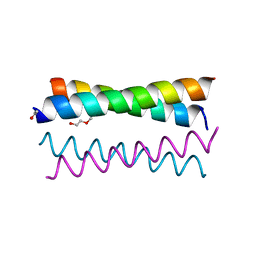 | | Crystal Structure of the de novo designed peptide alpha4H | | Descriptor: | ACETYL GROUP, TRIETHYLENE GLYCOL, alpha4H | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWG
 
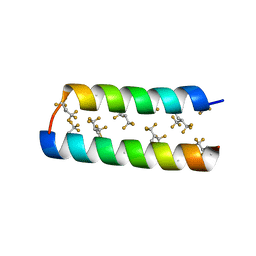 | |
4KVR
 
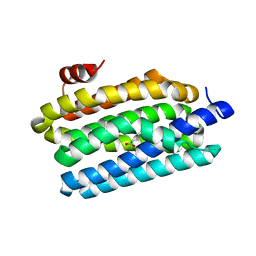 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVS
 
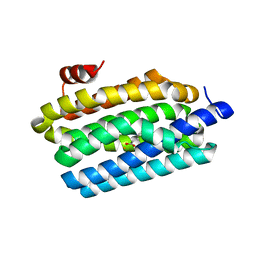 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVQ
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase wild type with palmitic acid bound | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, PALMITIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4PGK
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, FE (III) ION | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PG0
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PG1
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PGI
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Analogs Bound | | Descriptor: | 1,2-ETHANEDIOL, 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
