6O9A
 
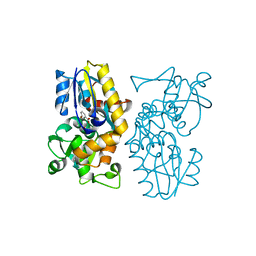 | | Crystal structure of MqnA complexed with 3-hydroxybenzoic acid | | Descriptor: | 3-HYDROXYBENZOIC ACID, ACETATE ION, Chorismate dehydratase | | Authors: | Hicks, K.A, Mahanta, N, Naseem, S, Fedoseyenko, D, Begley, T.P, Ealick, S.E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Menaquinone Biosynthesis: Biochemical and Structural Studies of Chorismate Dehydratase.
Biochemistry, 58, 2019
|
|
3O15
 
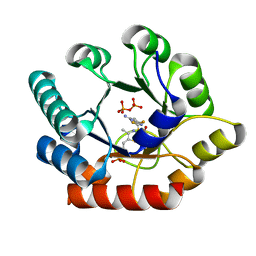 | | Crystal Structure of Bacillus subtilis Thiamin Phosphate Synthase Complexed with a Carboxylated Thiazole Phosphate | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, PYROPHOSPHATE 2-, ... | | Authors: | McCulloch, K.M, Hanes, J.W, Abdelwahed, S, Mahanta, N, Hazra, A, Ishida, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-07-20 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Kinetic Characterization of Bacillus subtilis
Thiamin Phosphate Synthase with a Carboxylated Thiazole Phosphate
to be published
|
|
3O16
 
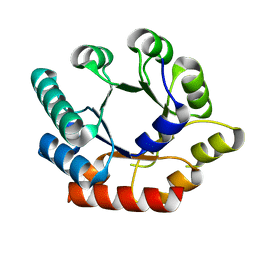 | | Crystal Structure of Bacillus subtilis Thiamin Phosphate Synthase K159A | | Descriptor: | Thiamine-phosphate pyrophosphorylase | | Authors: | McCulloch, K.M, Hanes, J.W, Abdelwahed, S, Mahanta, N, Hazra, A, Ishida, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-07-20 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Kinetic Characterization of Bacillus subtilis Thiamin
Phosphate Synthase with a Carboxylated Thiazole Phosphate
to be published
|
|
4GIL
 
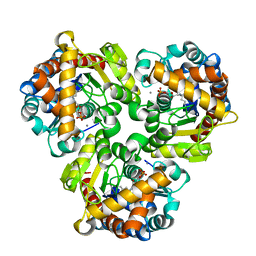 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase/Linear Pseudouridine 5'-Phosphate Adduct | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, pseudouridine 5'-phosphate, ... | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.539 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIJ
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Sulfate | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, SULFATE ION | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIM
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Pseudouridine 5'-phosphate | | Descriptor: | MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, Pseudouridine-5'-phosphate glycosidase | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4GIK
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase/Linear R5P Adduct | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, RIBOSE-5-PHOSPHATE | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
6CI7
 
 | |
6CIB
 
 | |
6XP8
 
 | |
6PE3
 
 | |
6PEU
 
 | |
