5WBF
 
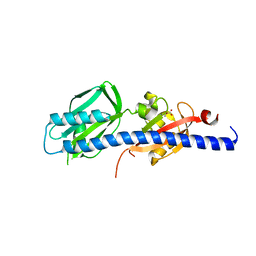 | | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | | Descriptor: | GLYCEROL, LACTIC ACID, Methyl-accepting chemotaxis transducer (TlpC) | | Authors: | Machuca, M.A, Johnson, K.S, Liu, Y.C, Steer, D.L, Ottemann, K.M, Roujeinikova, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate.
Sci Rep, 7, 2017
|
|
4WY9
 
 | |
6OFQ
 
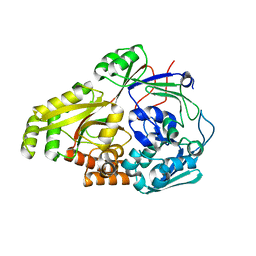 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori in complex with peptide STSA | | Descriptor: | Heme-binding protein A / AI-2 binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-03-31 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
6PU3
 
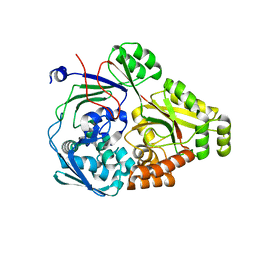 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori | | Descriptor: | Heme-binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
4XMR
 
 | | Crystal structure of the sensory domain of the Campylobacter jejuni chemoreceptor Tlp3 (CcmL) with isoleucine bound. | | Descriptor: | ISOLEUCINE, Putative methyl-accepting chemotaxis signal transduction protein, SULFATE ION | | Authors: | Roujeinikova, A, Liu, Y.C, Machuca, M.A. | | Deposit date: | 2015-01-15 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XMQ
 
 | |
6W3R
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 3-methylisoleucine | | Descriptor: | 3-methyl-L-alloisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3O
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 4-methylisoleucine | | Descriptor: | 4-methylisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3X
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-valine | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, SULFATE ION, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3P
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with beta-methylnorleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3T
 
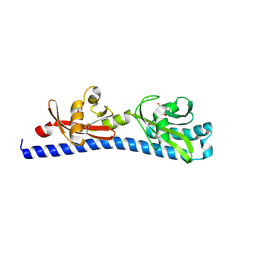 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-norvaline | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, NORVALINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3Y
 
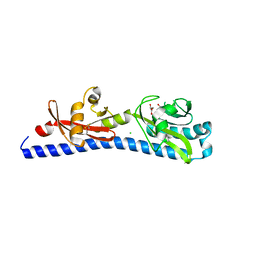 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-alanine | | Descriptor: | ALANINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3V
 
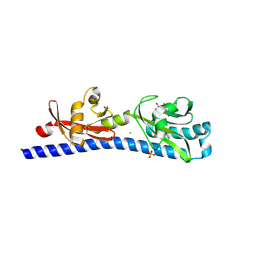 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-phenylalanine | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, PHENYLALANINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W3S
 
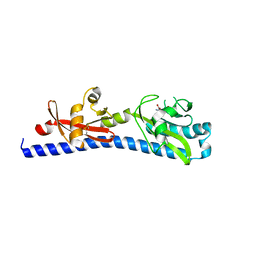 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-leucine | | Descriptor: | GLYCEROL, LEUCINE, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
4YHA
 
 | |
4YGF
 
 | | Crystal structure of the complex of Helicobacter pylori alpha-Carbonic Anhydrase with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Alpha-carbonic anhydrase, CHLORIDE ION, ... | | Authors: | Roujeinikova, A, Modak, J.K. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase by Sulfonamides.
Plos One, 10, 2015
|
|
