4QKR
 
 | |
4QKS
 
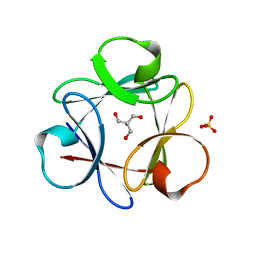 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | Authors: | Longo, L.M, Tenorio, C.A, Blaber, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
4OW4
 
 | |
4D8H
 
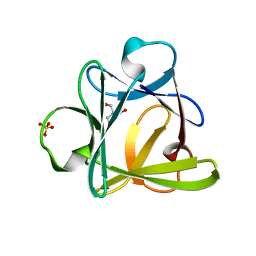 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | Authors: | Blaber, M, Longo, L. | | Deposit date: | 2012-01-10 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q7Y
 
 | |
3Q7W
 
 | |
3Q7X
 
 | | Crystal structure of Symfoil-4P/PV1: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo designed beta-trefoil architecture with symmetric primary structure | | Authors: | Blaber, M, Lee, J. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3BAG
 
 | |
3BAU
 
 | |
3BAQ
 
 | |
3BA5
 
 | |
3BAD
 
 | |
3BA4
 
 | |
3BAH
 
 | |
3BAO
 
 | |
3BA7
 
 | |
3BAV
 
 | |
3BB2
 
 | |
3B9U
 
 | |
4QAL
 
 | |
4Q9G
 
 | |
4QBC
 
 | |
4QBV
 
 | |
4Q9P
 
 | |
4QC4
 
 | |
