9F2K
 
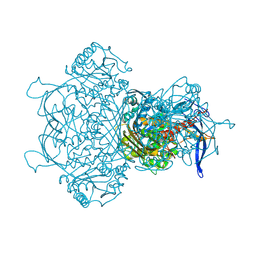 | | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, inositol-3-phosphate synthase | | Authors: | Traeger, T.K, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Disorder-to-order active site capping regulates the rate-limiting step of the inositol pathway.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OIU
 
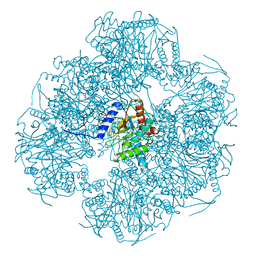 | | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Skalidis, I, Tueting, C, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon.
Commun Biol, 6, 2023
|
|
8PE8
 
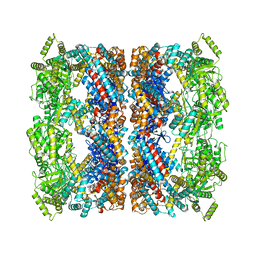 | |
7Q5Q
 
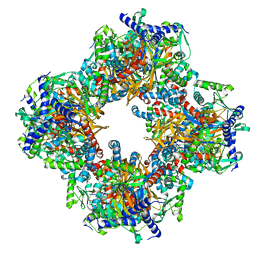 | | Protein community member oxoglutarate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5S
 
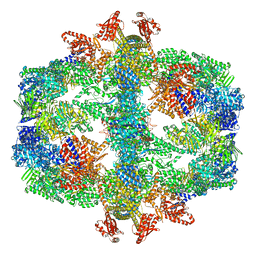 | | Protein community member fatty acid synthase complex from C. thermophilum | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5R
 
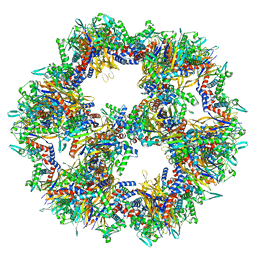 | | Protein community member pyruvate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7BGJ
 
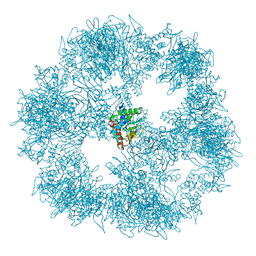 | | C. thermophilum Pyruvate Dehydrogenase Complex Core | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Tueting, C, Kastritis, P.L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts.
Cell Rep, 34, 2021
|
|
8A5T
 
 | |
6SHT
 
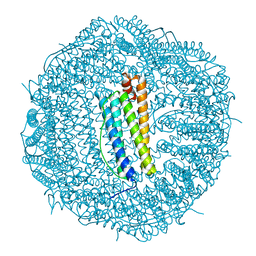 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
8C29
 
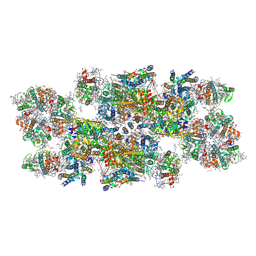 | | Cryo-EM structure of photosystem II C2S2 supercomplex from Norway spruce (Picea abies) at 2.8 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Kopecny, D, Semchonok, D.A, Kouril, R. | | Deposit date: | 2022-12-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.785 Å) | | Cite: | Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and alpha-tocopherol.
Nat.Plants, 9, 2023
|
|
7OTT
 
 | |
8PE4
 
 | |
8AE6
 
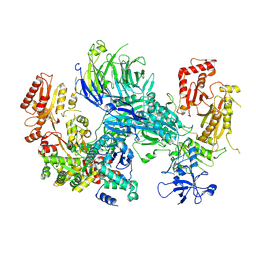 | | Cryo-EM structure of the SEA complex wing (SEACIT) | | Descriptor: | Maintenance of telomere capping protein 5, Nitrogen permease regulator 2, Nitrogen permease regulator 3, ... | | Authors: | Tafur, L, Loewith, R. | | Deposit date: | 2022-07-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the SEA complex.
Nature, 611, 2022
|
|
8ADL
 
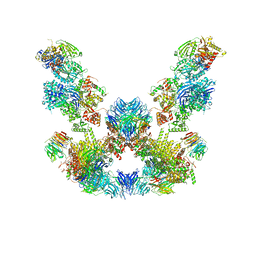 | | Cryo-EM structure of the SEA complex | | Descriptor: | Maintenance of telomere capping protein 5, Nitrogen permease regulator 2, Nitrogen permease regulator 3, ... | | Authors: | Tafur, L, Loewith, R. | | Deposit date: | 2022-07-08 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of the SEA complex.
Nature, 611, 2022
|
|
