1A2D
 
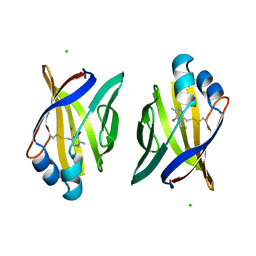 | | PYRIDOXAMINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN, CHLORIDE ION | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-29 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
1A18
 
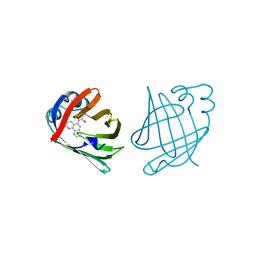 | | PHENANTHROLINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-23 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
9B3L
 
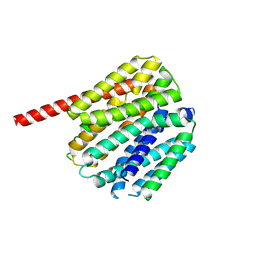 | |
9B3K
 
 | | NorA in complex with Fab36 (NorA-BRIL fusion) | | Descriptor: | Fab36 heavy chain, Fab36 light chain, NorA-BRIL(3A) fusion | | Authors: | Xie, P, Li, Y, Kuang, H, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A shared helix approach to solve small membrane transporter structures using cryo-EM
To Be Published
|
|
9B3M
 
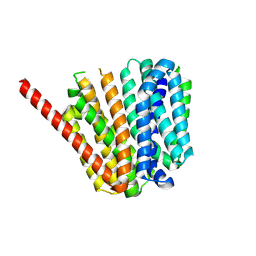 | |
9B3O
 
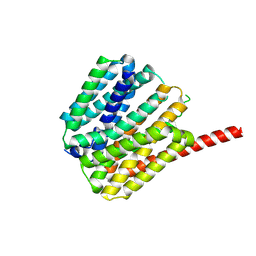 | |
8UVB
 
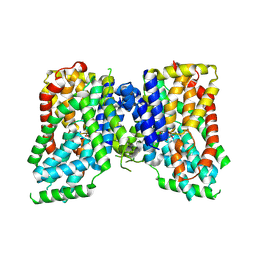 | | Structure of NaCT-PF4a complex | | Descriptor: | (2R)-2-hydroxy-2-[2-(2-methoxy-5-methylpyridin-3-yl)ethyl]butanedioic acid, SODIUM ION, Solute carrier family 13 member 5 | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVC
 
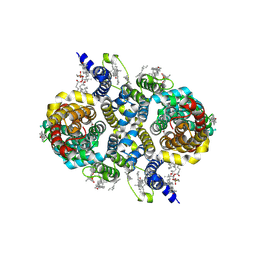 | | Structure of NaDC3-aKG complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-OXOGLUTARIC ACID, CHOLESTEROL, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVE
 
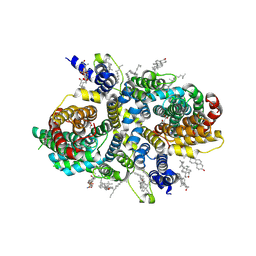 | | Structure of NaDC3-Succinate complex in Coo-Ci conformation | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, Na(+)/dicarboxylate cotransporter 3, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVG
 
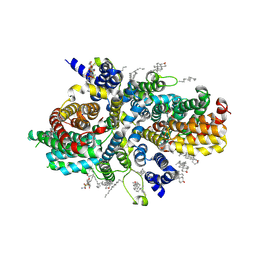 | | Structure of NaDC3-DMS complex in Co-Ci conformation | | Descriptor: | (2R,3S)-2,3-dimethylbutanedioic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVH
 
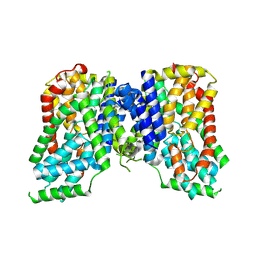 | | Structure of NaCT-succ complex | | Descriptor: | Solute carrier family 13 member 5 | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVI
 
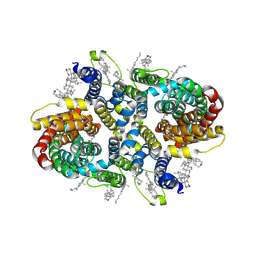 | | Structure of NaDC3-Succinate complex in Coo-Coo conformation | | Descriptor: | CHOLESTEROL, Na(+)/dicarboxylate cotransporter 3, SODIUM ION, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-03 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVF
 
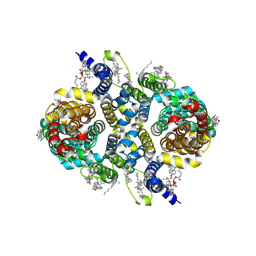 | | Structure of NaDC3-DMS complex in Ci-Ci conformation | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, Na(+)/dicarboxylate cotransporter 3, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8UVD
 
 | | Structure of NaDC3-PF4a complex | | Descriptor: | (2R)-2-hydroxy-2-[2-(2-methoxy-5-methylpyridin-3-yl)ethyl]butanedioic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, ... | | Authors: | Li, Y, Wang, D.N, Mindell, J.A, Rice, W.J, Song, J, Mikusevic, V, Marden, J.J, Becerril, A, Kuang, H, Wang, B. | | Deposit date: | 2023-11-02 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8TTH
 
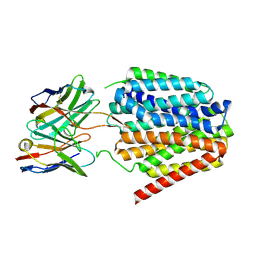 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTF
 
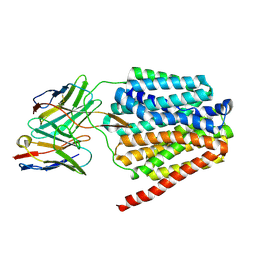 | | NorA double mutant - E222QD307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTE
 
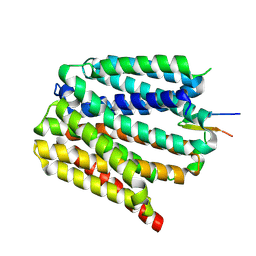 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTG
 
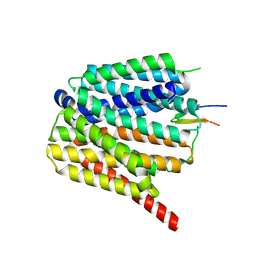 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
7LTO
 
 | | Nse5-6 complex | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Li, S.B, Zheng, S, Tangy, S, Koyi, C, Wan, B.B, Kung, H.H, Andrej, S, Alex, K, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Integrative analysis reveals unique structural and functional features of the Smc5/6 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7TVE
 
 | | ATP and DNA bound SMC5/6 core complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (68-MER), DNA (78-MER), ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9CEY
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 5 | | Descriptor: | DNA (26-MER), DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF3
 
 | | Parasitella parasitica Fanzor (PpFz) State 4 | | Descriptor: | DNA non-target strand, DNA target strand, MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CER
 
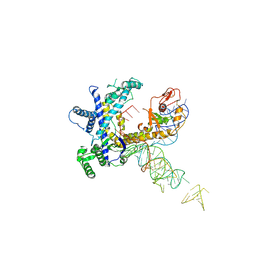 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEU
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 1 | | Descriptor: | DNA (5'-D(P*CP*CP*TP*AP*TP*AP*GP*AP*TP*AP*TP*GP*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), Maltose/maltodextrin-binding periplasmic protein,Spizellomyces punctatus Fanzor 1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEV
 
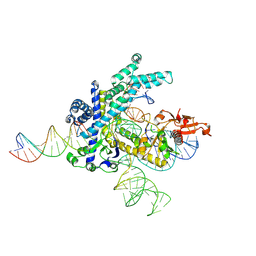 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
