5IP3
 
 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssDNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
5IP2
 
 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssRNA complex | | Descriptor: | Nucleoprotein, RNA (5'-D(P*UP*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
5IP1
 
 | | Tomato spotted wilt tospovirus nucleocapsid protein | | Descriptor: | Nucleoprotein | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
4I86
 
 | | Crystal structure of PilZ domain of CeSA from cellulose synthesizing bacterium | | Descriptor: | Cellulose synthase 1 | | Authors: | Fujiwara, T, Komoda, K, Sakurai, N, Tanaka, I, Yao, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A
Biochem.Biophys.Res.Commun., 431, 2013
|
|
4XD9
 
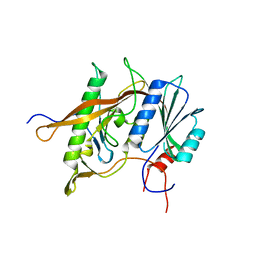 | | Structure of Rpf2-Rrs1 complex involved in ribosome biogenesis | | Descriptor: | Ribosome biogenesis protein (Rrs1), putative (AFU_orthologue AFUA_7G04430), Ribosome biogenesis protein, ... | | Authors: | Asano, N, Kato, K, Yao, M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis.
Nucleic Acids Res., 43, 2015
|
|
5AYE
 
 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate and beta-(1,4)-mannobiose | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5AYD
 
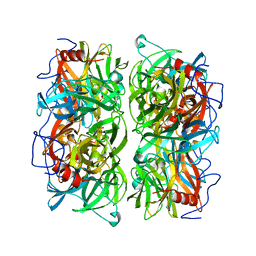 | | Crystal structure of Ruminococcus albus beta-(1,4)-mannooligosaccharide phosphorylase (RaMP2) in complexes with phosphate | | Descriptor: | Beta-1,4-mannooligosaccharide phosphorylase, PHOSPHATE ION | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
5AY9
 
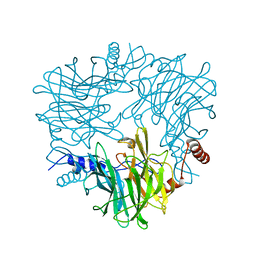 | |
5AYC
 
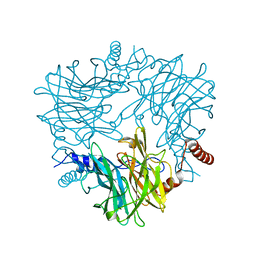 | | Crystal structure of Ruminococcus albus 4-O-beta-D-mannosyl-D-glucose phosphorylase (RaMP1) in complexes with sulfate and 4-O-beta-D-mannosyl-D-glucose | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, SULFATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ye, Y, Saburi, W, Kato, K, Yao, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies.
Febs Lett., 590, 2016
|
|
3WC0
 
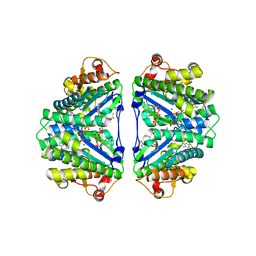 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Likely histidyl tRNA-specific guanylyltransferase, MAGNESIUM ION | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WC1
 
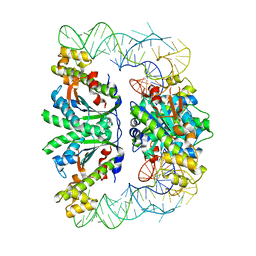 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a G-1 deleted tRNA(His) | | Descriptor: | 75-mer tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WBZ
 
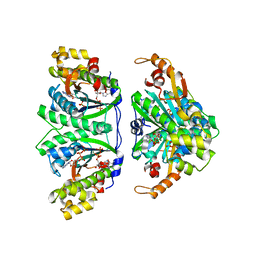 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Likely histidyl tRNA-specific guanylyltransferase, MAGNESIUM ION | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WC2
 
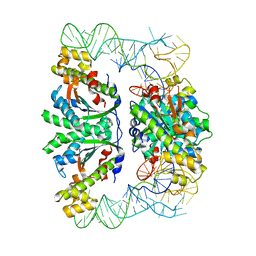 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a tRNA(Phe)(GUG) | | Descriptor: | 76mer-tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.641 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WY2
 
 | | Crystal structure of alpha-glucosidase in complex with glucose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY1
 
 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY3
 
 | | Crystal structure of alpha-glucosidase mutant D202N in complex with glucose and glycerol | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY4
 
 | | Crystal structure of alpha-glucosidase mutant E271Q in complex with maltose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
