1SUH
 
 | | AMINO-TERMINAL DOMAIN OF EPITHELIAL CADHERIN IN THE CALCIUM BOUND STATE, NMR, 20 STRUCTURES | | Descriptor: | EPITHELIAL CADHERIN | | Authors: | Overduin, M, Tong, K.I, Kay, C.M, Ikura, M. | | Deposit date: | 1996-01-30 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and monomeric structure of the amino-terminal extracellular domain of epithelial cadherin.
J.Biomol.NMR, 7, 1996
|
|
1PAK
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1PAJ
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
2A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, 40 STRUCTURES | | Descriptor: | C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-06-09 | | Release date: | 1999-01-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
1A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-04-15 | | Release date: | 1998-10-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
8U41
 
 | |
8U42
 
 | |
8UX5
 
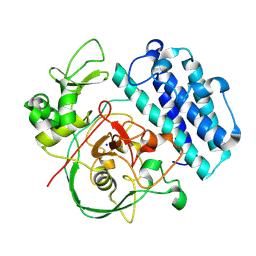 | |
1KB8
 
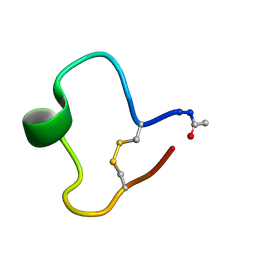 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1TNX
 
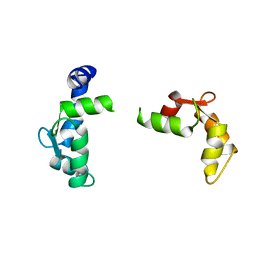 | |
1TNW
 
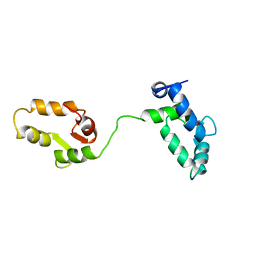 | |
1KB7
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | KB7 PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1NIL
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAK PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1NIM
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAK PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAN
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PAO
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAO PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1PON
 
 | |
1IKM
 
 | |
1IKL
 
 | |
1DVI
 
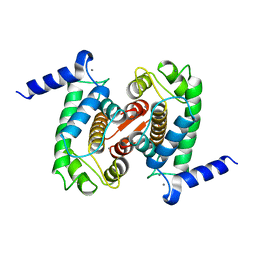 | | CALPAIN DOMAIN VI WITH CALCIUM BOUND | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Cygler, M, Blanchard, H, Grochulski, P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
1AJ5
 
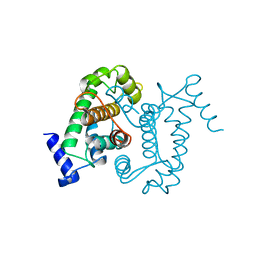 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
8FBX
 
 | | Selenosugar synthase SenB from Variovorax paradoxus | | Descriptor: | CHLORIDE ION, SODIUM ION, Selenosugar synthase SenB | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Characterization and Ligand-Induced Conformational Changes of SenB, a Se-Glycosyltransferase Involved in Selenoneine Biosynthesis.
Biochemistry, 62, 2023
|
|
7MER
 
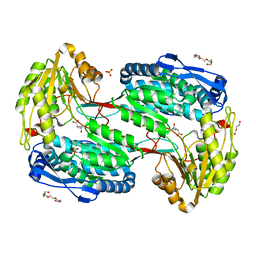 | | Structure of ALDH4A1 complexed with trans-4-Hydroxy-L-proline | | Descriptor: | 4-HYDROXYPROLINE, DI(HYDROXYETHYL)ETHER, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Bogner, A.N, Stiers, K.M, Tanner, J.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the stereospecific inhibition of the dual proline/hydroxyproline catabolic enzyme ALDH4A1 by trans-4-hydroxy-L-proline.
Protein Sci., 30, 2021
|
|
7MES
 
 | | Structure of ALDH4A1 complexed with trans-4-Hydroxy-D-proline | | Descriptor: | (4S)-4-hydroxy-D-proline, DI(HYDROXYETHYL)ETHER, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Bogner, A.N, Stiers, K.M, Tanner, J.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for the stereospecific inhibition of the dual proline/hydroxyproline catabolic enzyme ALDH4A1 by trans-4-hydroxy-L-proline.
Protein Sci., 30, 2021
|
|
