4MGJ
 
 | | Crystal structure of cytochrome P450 2B4 F429H in complex with 4-CPI | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, Y, Zhang, H, Usharani, D, Bu, W, Im, S, Tarasev, M, Rwere, F, Meagher, J, Sun, C, Stuckey, J, Shaik, S, Waskell, L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Characterization of a Cytochrome P450 2B4 F429H Mutant with an Axial Thiolate-Histidine Hydrogen Bond.
Biochemistry, 53, 2014
|
|
2M33
 
 | | Solution NMR structure of full-length oxidized microsomal rabbit cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Subramanian, V, Ahuja, S, Popovych, N, Huang, R, Le Clair, S.V, Jahr, N, Soong, R, Xu, J, Yamamoto, K, Nanga, R.P, Im, S, Waskell, L, Ramamoorthy, A. | | Deposit date: | 2013-01-08 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of full-length mammalian cytochrome b5
To be Published
|
|
8WFC
 
 | |
2GZB
 
 | | Bauhinia bauhinioides cruzipain inhibitor (BbCI) | | Descriptor: | IODIDE ION, Kunitz-type proteinase inhibitor BbCI | | Authors: | Hansen, D, Macedo-Ribeiro, S, Navarro, M.V.A.S, Garratt, R.C, Oliva, M.L.V. | | Deposit date: | 2006-05-11 | | Release date: | 2007-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a novel cysteinless plant Kunitz-type protease inhibitor.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
3IOS
 
 | | Structure of MTB dsbF in its mixed oxidized and reduced forms | | Descriptor: | Disulfide bond forming protein (DsbF) | | Authors: | Chim, N, Goulding, C.W. | | Deposit date: | 2009-08-14 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An extracellular disulfide bond forming protein (DsbF) from Mycobacterium tuberculosis: structural, biochemical, and gene expression analysis.
J.Mol.Biol., 396, 2010
|
|
6BWW
 
 | |
9ISP
 
 | | DUF2436 domain which is frequently found in virulence proteins from Porphyromonas gingivalis | | Descriptor: | Protease PrtH | | Authors: | Kim, B, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2024-07-18 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | First crystal structure of the DUF2436 domain of virulence proteins from Porphyromonas gingivalis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
5URH
 
 | | CYPOR/D632A with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URD
 
 | | wild type rat CYPOR bound with NADP+ - oxidized form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URI
 
 | | Rat CYPOR/D632A with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URE
 
 | | Wild type rat CYPOR bound with NADP+ - reduced form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URG
 
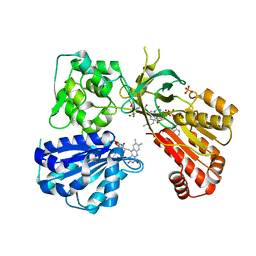 | | rat CYPOR D632F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
4Y9R
 
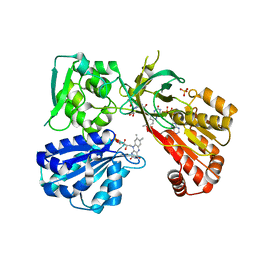 | | rat CYPOR mutant - G141del | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAL
 
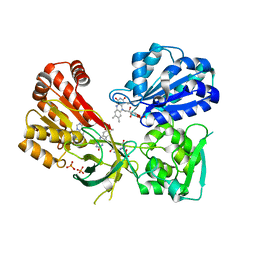 | | Reduced CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAO
 
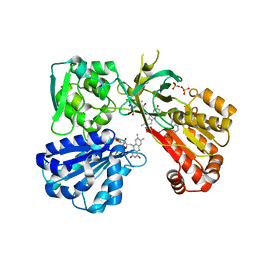 | | Reduced CYPOR mutant - G143del | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAW
 
 | | Reduced CYPOR mutant - G141del | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAU
 
 | | Reduced CYPOR mutant - G141del/E142N | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4Y9U
 
 | | rat CYPOR mutant - G143del | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4YAF
 
 | | rat CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4Y7C
 
 | | rat CYPOR mutant - G141del/E142N | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-13 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
