1AXU
 
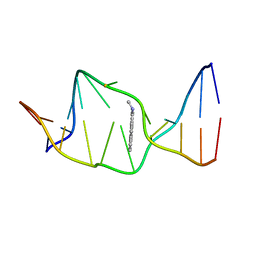 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
1C0Y
 
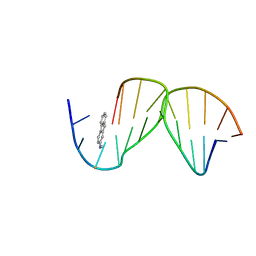 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1XKT
 
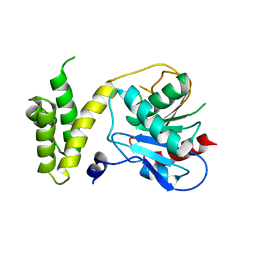 | | Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain | | Descriptor: | fatty acid synthase | | Authors: | Chakravarty, B, Gu, Z, Chirala, S.S, Wakil, S.J, Quiocho, F.A. | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1AX6
 
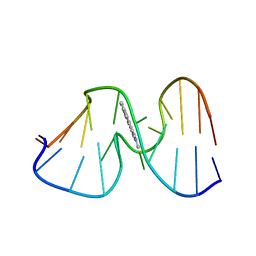 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT OPPOSITE A-2 DELETION SITE IN THE NARI HOT SPOT SEQUENCE CONTEXT; NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(CTCGGC-[AF]G-CCATC)D(GATGGCCGAG) | | Authors: | Mao, B, Gorin, A.A, Gu, Z, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a--2 deletion site in the NarI hot spot sequence context.
Biochemistry, 36, 1997
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AX7
 
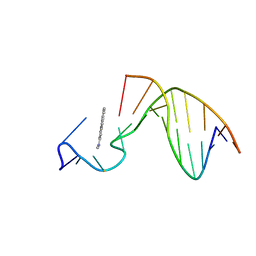 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(AAC-[AF]G-CTACCATCC)D(GGATGGTAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction.
Biochemistry, 36, 1997
|
|
7WOL
 
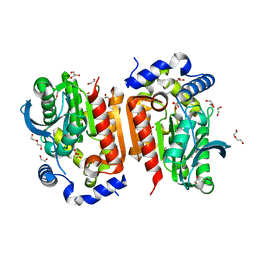 | | Crystal structure of lipase TrLipB from Thermomocrobium roseum | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, L, Fang, Y, Shi, Y, Gu, Z, Xin, Y. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lipase TrLipB from Thermomocrobium roseum
To Be Published
|
|
8XOU
 
 | | Prohead portal vertex of bacteriophage lambda | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XQB
 
 | | Mature virion portal vertex of bacteriophage lambda | | Descriptor: | Capsid decoration protein, Head completion protein, Head-tail connector protein FII, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XPM
 
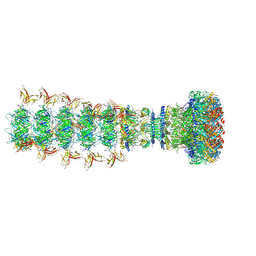 | | Mature virion portal of phage lambda with DNA | | Descriptor: | DNA (104-MER), DNA (92-MER), Head completion protein, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XOT
 
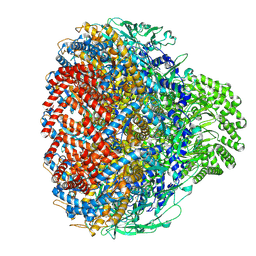 | | Prohead portal of bacteriophage lambda | | Descriptor: | Portal protein B | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XOW
 
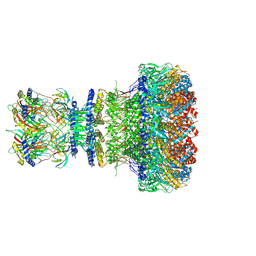 | | Mature virion portal of bacteriophage lambda | | Descriptor: | Head completion protein, Head-tail connector protein FII, Portal protein B, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6AG0
 
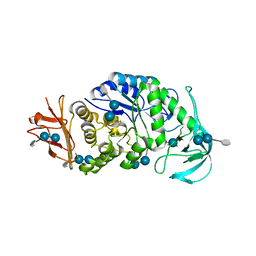 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
6INM
 
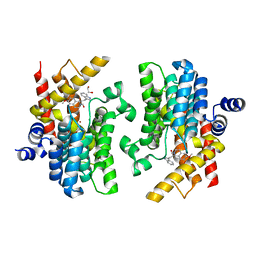 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMB
 
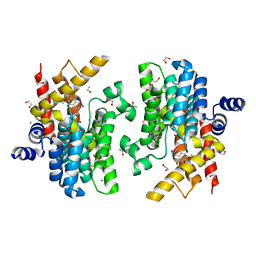 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMI
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-ethoxy-7-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMR
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[3-(1H-indol-3-yl)propyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6INK
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMD
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IND
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-6,7-dimethoxy-1-[2-(6-methyl-1H-indol-3-yl)ethyl]-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMO
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[(1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IM6
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMT
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(6-fluoro-1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
