8QY4
 
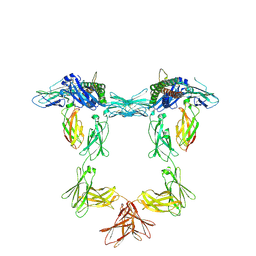 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY5
 
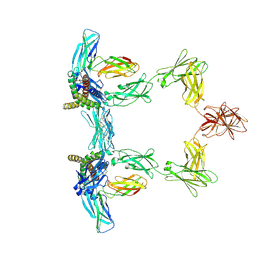 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8QY6
 
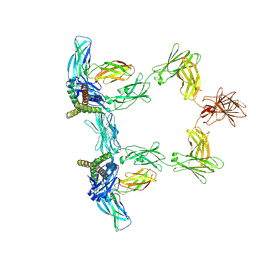 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into IL-11-mediated signalling and human IL6ST variant-associated immunodeficiency.
Nat Commun, 15, 2024
|
|
8BA7
 
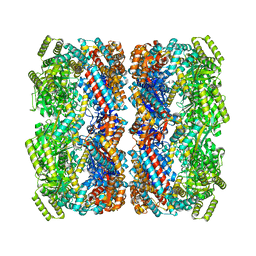 | |
8BA8
 
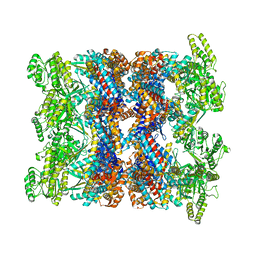 | | CryoEM structure of GroEL-ADP.BeF3-Rubisco. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BA9
 
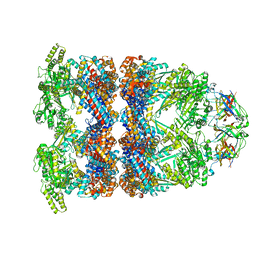 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
7Z0L
 
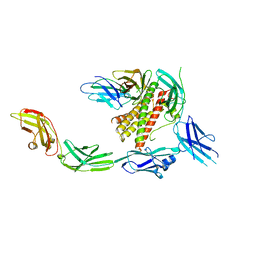 | | IL-27 signalling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Jin, Y, Gardner, S, Bubeck, D. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the assembly and activation of the IL-27 signaling complex.
Embo Rep., 23, 2022
|
|
6UL7
 
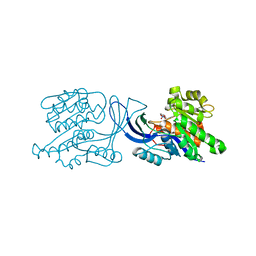 | | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Gardner, S, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-10-07 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole
To be Published
|
|
8B0H
 
 | | 2C9, C5b9-CD59 cryoEM structure | | Descriptor: | CD59 glycoprotein, Complement C5, Complement component C6, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bubeck, D, Couves, E.C, Gardner, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
6H03
 
 | | OPEN CONFORMATION OF THE MEMBRANE ATTACK COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
6H04
 
 | | Closed conformation of the Membrane Attack Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
8G0L
 
 | |
7KJE
 
 | | F96S epi-isozizaene synthase: complex with 3 Mg2+ and neridronate | | Descriptor: | (6-azanyl-1-oxidanyl-1-phosphono-hexyl)phosphonic acid, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ronnebaum, T.A, Gardner, S, Christianson, D.W. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Biochemistry, 59, 2020
|
|
6P2D
 
 | | Structure of mouse ketohexokinase-C in complex with fructose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Michaelis-like complex of mouse ketohexokinase isoform C
ACTA CRYSTALLOGR.,SECT.D, 2024
|
|
8C6I
 
 | | TMEM2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hohenester, E. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the transmembrane protein 2 (TMEM2) ectodomain and its apparent lack of hyaluronidase activity.
Wellcome Open Res, 8, 2023
|
|
