2W0N
 
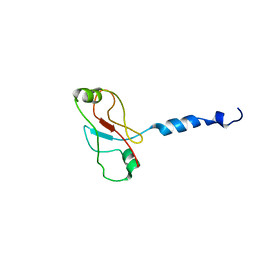 | | Plasticity of PAS domain and potential role for signal transduction in the histidine-kinase DcuS | | Descriptor: | SENSOR PROTEIN DCUS | | Authors: | Etzkorn, M, Kneuper, H, Duennwald, P, Vijayan, V, Kraemer, J, Griesinger, C, Becker, S, Unden, G, Baldus, M. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Plasticity of the Pas Domain and a Potential Role for Signal Transduction in the Histidine Kinase Dcus.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7PDU
 
 | |
6GSF
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
5OVM
 
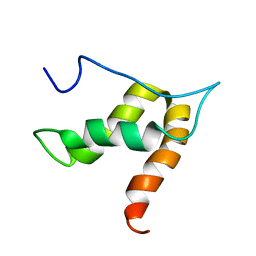 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2017-08-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M06
 
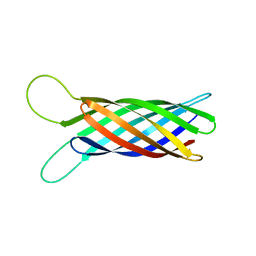 | | NMR structure of OmpX in phopspholipid nanodiscs | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
6SR7
 
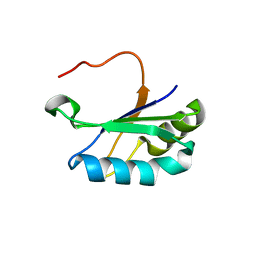 | | Structure of the U1A variant A1-98 Y31H/Q36R/K98W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
6SQQ
 
 | |
6SQV
 
 | | Structure of the U1A variant A1-98 Y31H/Q36R/R70W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
6SQT
 
 | |
6SQN
 
 | |
