6GSC
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, T.D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
6ZGP
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with inhibitor MMV12 (MMV020670) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D.H, Cameron, A, Chen, Y. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
1U2E
 
 | | Crystal Structure of the C-C bond hydrolase MhpC | | Descriptor: | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase, CHLORIDE ION | | Authors: | Montgomery, M.G, Dunn, G, Mohammed, F, Robertson, T, Garcia, J.-L, Coker, A, Bugg, T.D.H, Wood, S.P. | | Deposit date: | 2004-07-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the C-C Bond Hydrolase MhpC Provides Insights into its Catalytic Mechanism
J.Mol.Biol., 346, 2005
|
|
5FW4
 
 | | Structure of Thermobifida fusca DyP-type Peroxidase and Activity towards Kraft Lignin and Lignin Model Compounds | | Descriptor: | DYE-DECOLORIZING PEROXIDASE TFU_3078, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rahmanpour, R, Rea, D, Jamshidi, S, Fulop, V, Bugg, T.D.H. | | Deposit date: | 2016-02-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermobifida Fusca Dyp-Type Peroxidase and Activity Towards Kraft Lignin and Lignin Model Compounds.
Arch.Biochem.Biophys., 594, 2016
|
|
5A9G
 
 | | Manganese Superoxide Dismutase from Sphingobacterium sp. T2 | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Rashid, G.M.M, Taylor, C.R, Liu, Y, Zhang, X, Rea, D, Fulop, V, Bugg, T.D.H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Manganese Superoxide Dismutase from Sphingobacterium Sp. T2 as a Novel Bacterial Enzyme for Lignin Oxidation.
Acs Chem.Biol., 10, 2015
|
|
2VWS
 
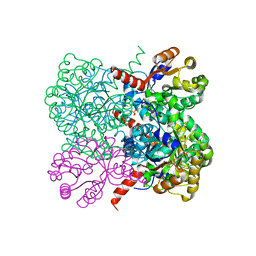 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 | | Descriptor: | GLYCEROL, PHOSPHATE ION, YFAU, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
2VWT
 
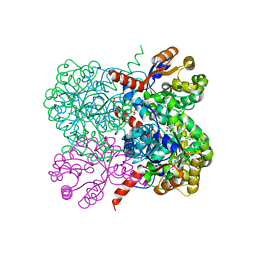 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
2V5J
 
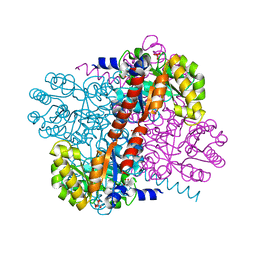 | | Apo Class II aldolase HpcH | | Descriptor: | 2,4-DIHYDROXYHEPT-2-ENE-1,7-DIOIC ACID ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Rea, D, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2007-07-05 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of Hpch: A Metal Ion Dependent Class II Aldolase from the Homoprotocatechuate Degradation Pathway of Escherichia Coli.
J.Mol.Biol., 373, 2007
|
|
2V5K
 
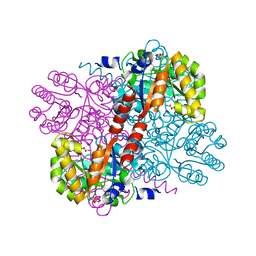 | | Class II aldolase HpcH - magnesium - oxamate complex | | Descriptor: | 2,4-DIHYDROXYHEPT-2-ENE-1,7-DIOIC ACID ALDOLASE, MAGNESIUM ION, OXAMIC ACID, ... | | Authors: | Rea, D, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2007-07-06 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of Hpch: A Metal Ion Dependent Class II Aldolase from the Homoprotocatechuate Degradation Pathway of Escherichia Coli.
J.Mol.Biol., 373, 2007
|
|
6GSB
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
3S2U
 
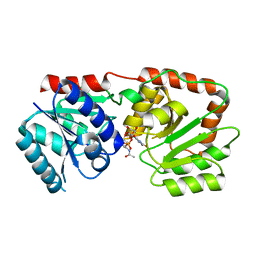 | | Crystal structure of the Pseudomonas aeruginosa MurG:UDP-GlcNAc substrate complex | | Descriptor: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Brown, K, Vial, S.C.M, Dedi, N, Westcott, J, Scally, S, Bugg, T.D.H, Charlton, P.A, Cheetham, G.M.T. | | Deposit date: | 2011-05-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa MurG: UDP-GlcNAc Substrate Complex.
Protein Pept.Lett., 20, 2013
|
|
6Y8J
 
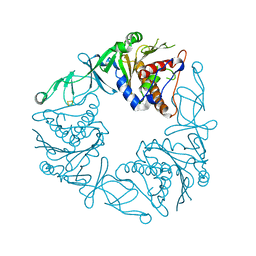 | | Crystal structure of the apo form of a quaternary ammonium Rieske monooxygenase CntA | | Descriptor: | Carnitine monooxygenase oxygenase subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y9D
 
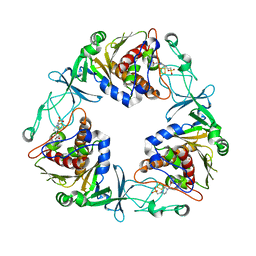 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate L-Carnitine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARNITINE, Carnitine monooxygenase oxygenase subunit, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y8S
 
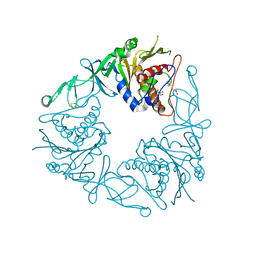 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y9C
 
 | | The structure of a quaternary ammonium Rieske monooxygenase reveals insights into carnitine oxidation by gut microbiota and inter-subunit electron transfer | | Descriptor: | CARNITINE, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterisation of an unusual cysteine pair in the Rieske carnitine monooxygenase CntA catalytic site.
Febs J., 2023
|
|
6QSI
 
 | |
7ORX
 
 | |
6TJ2
 
 | |
6EVG
 
 | |
