1QMT
 
 | |
4A2O
 
 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH SULFATE ANIONS | | Descriptor: | EOSINOPHIL CATIONIC PROTEIN, SULFATE ION | | Authors: | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
4A2Y
 
 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH CITRATE ANIONS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
1GX4
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - N-ACETYL LACTOSAMINE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-27 | | Release date: | 2003-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GWW
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GX0
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - BETA-D-GALACTOSE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GWV
 
 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - LACTOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1K4V
 
 | | 1.53 A Crystal Structure of the Beta-Galactoside-alpha-1,3-galactosyltransferase in Complex with UDP | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Swaminathan, G.J, Zhang, Y, Natesh, R, Brew, K, Acharya, K.R. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of UDP complex of UDP-galactose:beta-galactoside-alpha -1,3-galactosyltransferase at 1.53-A resolution reveals a conformational change in the catalytically important C terminus.
J.Biol.Chem., 276, 2001
|
|
6ENP
 
 | | Atomic resolution structure of human RNase 6 in the presence of phosphate anions in P21 space group. | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Ribonuclease K6, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.042 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
3KGQ
 
 | | Carboxypeptidase A liganded to an organic small-molecule: conformational changes | | Descriptor: | ACETONE, CITRIC ACID, Carboxypeptidase A1, ... | | Authors: | Fernandez, D, Boix, E, Pallares, I, Aviles, F.X, Vendrell, J. | | Deposit date: | 2009-10-29 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analysis of the Complex between Citrate and the Zinc Peptidase Carboxypeptidase A
Enzyme Res, 2011, 2011
|
|
8QEW
 
 | |
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
1HI3
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
5OAB
 
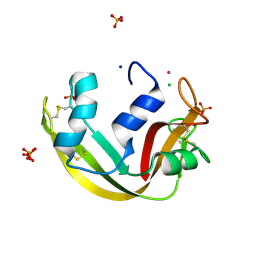 | | A novel crystal form of human RNase6 at atomic resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.111 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4OXB
 
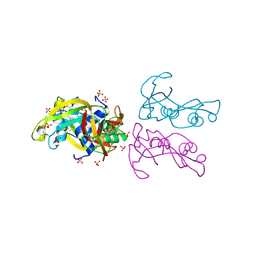 | |
4OXF
 
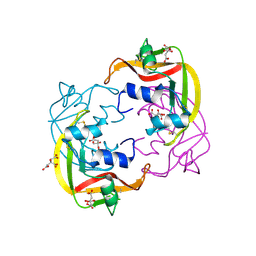 | |
4X09
 
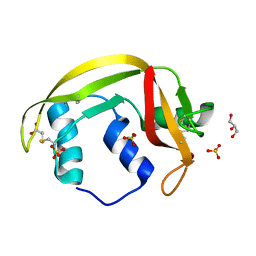 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
1H1H
 
 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | Deposit date: | 2002-07-15 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
5ET4
 
 | | Structure of RNase A-K7H/R10H in complex with 3'-CMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-3'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Blanco, J.A, Salazar, V.A, Moussaoui, M, Boix, E. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6SSN
 
 | | RNASE 3/1 version3 | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNase 3/1 version3 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6SSO
 
 | | EDN mutant L45H | | Descriptor: | ACETATE ION, Non-secretory ribonuclease | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and functional characterization of new family enzymes derivates from human RNase 1 and 3 with antimicrobial and ribonuclease activity
To Be Published
|
|
5OGH
 
 | | Structure of RNase A at high resolution (1.16 A) in complex with 3'-CMP and sulphate ions | | Descriptor: | CHLORIDE ION, CYTIDINE-3'-MONOPHOSPHATE, Ribonuclease pancreatic, ... | | Authors: | Blanco, J.A, Prats-Ejarque, G, Salazar, V.A, Moussaoui, M, Boix, E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6YBC
 
 | | RNASE 3/1 version2 phosphate complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, RNASE 3/1 version2 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-03-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6YMT
 
 | | RNASE 3/1 version1 | | Descriptor: | GLYCEROL, RNase 3/1 version 1 | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring the RNase A scaffold to combine catalytic and antimicrobial activities. Structural characterization of RNase 3/1 chimeras.
Front Mol Biosci, 9, 2022
|
|
6YBE
 
 | |
