3GBP
 
 | |
2CJP
 
 | | Structure of potato (Solanum tuberosum) epoxide hydrolase I (StEH1) | | Descriptor: | 1,2-ETHANEDIOL, 2-PROPYLPENTANAMIDE, EPOXIDE HYDROLASE, ... | | Authors: | Mowbray, S.L, Elfstrom, L.T, Ahlgren, K.M, Andersson, C.E, Widersten, M. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-Ray Structure of Potato Epoxide Hydrolase Sheds Light on Substrate Specificity in Plant Enzymes.
Protein Sci., 15, 2006
|
|
1DRJ
 
 | |
1DRK
 
 | |
1DBP
 
 | |
2DRI
 
 | |
5JAZ
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC51 and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, FORMIC ACID, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-12 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
1W74
 
 | | X-ray structure of peptidyl-prolyl cis-trans isomerase A, PpiA, Rv0009, from Mycobacterium tuberculosis. | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Henriksson, L.M, Johansson, P, Unge, T, Mowbray, S.L. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Peptidyl-Prolyl Cis-Trans Isomerase a from Mycobacterium Tuberculosis
Eur.J.Biochem., 271, 2004
|
|
1GCA
 
 | |
1GCG
 
 | |
3ZHX
 
 | | Structure of Mycobacterium tuberculosis DXR in complex with a fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, [(1S)-1-(3,4-dichlorophenyl)-3-[oxidanyl(phenylcarbonyl)amino]propyl]phosphonic acid | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
3ZI0
 
 | | Structure of Mycobacterium tuberculosis DXR in complex with a fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, [(1S)-1-(3,4-dichlorophenyl)-3-{hydroxy[2-(1H-1,2,4-triazol-1-ylmethyl)benzoyl]amino}propyl]phosphonic acid | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
3ZHY
 
 | | Structure of Mycobacterium tuberculosis DXR in complex with a di- substituted fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
3ZHZ
 
 | | Structure of Mycobacterium tuberculosis DXR in complex with a fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, [(1S)-1-(3,4-dichlorophenyl)-3-[oxidanyl-[2-[[3-(trifluoromethyl)phenyl]amino]phenyl]carbonyl-amino]propyl]phosphonic acid | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
5JMP
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC57 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5JMW
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC50 and manganese | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
1MPD
 
 | |
1MPB
 
 | |
1MPC
 
 | |
1BA2
 
 | |
1Z3W
 
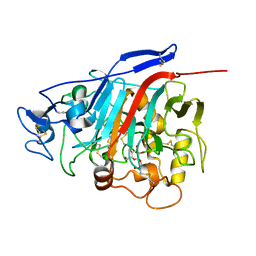 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Vasella, A, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3V
 
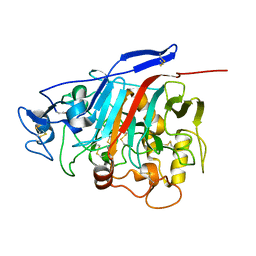 | | Structure of Phanerochaete chrysosporium cellobiohydrolase Cel7D (CBH58) in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, cellulase | | Authors: | Ubhayasekera, W, Munoz, I.G, Stahlberg, J, Mowbray, S.L. | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors
Febs J., 272, 2005
|
|
1Z3T
 
 | |
1GQT
 
 | |
3ZXR
 
 | |
