3GL1
 
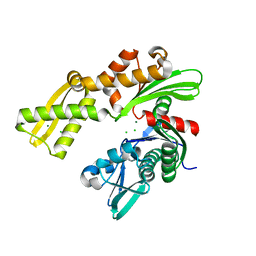 | | Crystal structure of ATPase domain of Ssb1 chaperone, a member of the HSP70 family, from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock protein SSB1, ... | | Authors: | Osipiuk, J, Li, H, Bargassa, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of ATPase domain of Ssb1 chaperone, member of the HSP70 family from Saccharomyces cerevisiae.
To be Published
|
|
3GSE
 
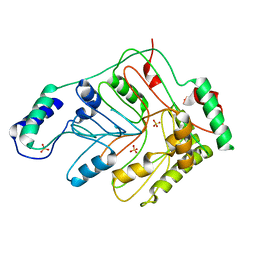 | | Crystal structure of menaquinone-specific isochorismate synthase from Yersinia pestis CO92 | | Descriptor: | Menaquinone-specific isochorismate synthase, SULFATE ION | | Authors: | Nocek, B, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-26 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of menaquinone-specific isochorismate synthase from Yersinia pestis CO92
To be Published
|
|
3LHF
 
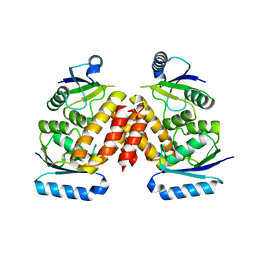 | | The Crystal Structure of a Serine Recombinase from Sulfolobus solfataricus to 2.3A | | Descriptor: | Serine Recombinase | | Authors: | Stein, A.J, Osipiuk, J, Marshall, N, Bearden, J, Davidoff, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-03-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of a Serine Recombinase from Sulfolobus solfataricus to 2.3A
To be Published
|
|
3LHQ
 
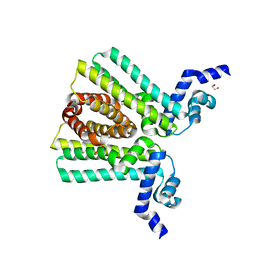 | | DNA-binding transcriptional repressor AcrR from Salmonella typhimurium. | | Descriptor: | 1,2-ETHANEDIOL, AcrAB operon repressor (TetR/AcrR family), DI(HYDROXYETHYL)ETHER | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of DNA-binding transcriptional repressor AcrR from Salmonella typhimurium.
To be Published
|
|
3L5Z
 
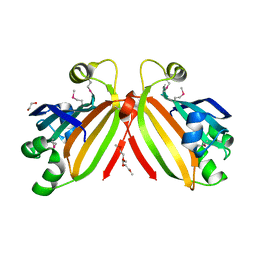 | | Crystal structure of transcriptional regulator, GntR family from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Transcriptional regulator, ... | | Authors: | Chang, C, Hatzos, C, Feldmann, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of transcriptional regulator, GntR family from Bacillus cereus
To be Published
|
|
3LHE
 
 | | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne | | Descriptor: | CHLORIDE ION, GLYCEROL, GntR family Transcriptional regulator | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne
To be Published
|
|
3LDU
 
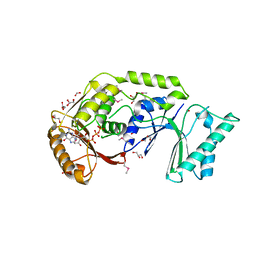 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3LHK
 
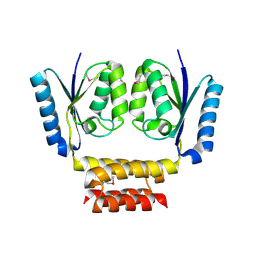 | |
3LJL
 
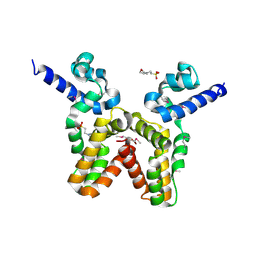 | |
3LEC
 
 | | The Crystal Structure of a protein in the NADB-Rossmann Superfamily from Streptococcus agalactiae to 1.8A | | Descriptor: | NADB-Rossmann Superfamily protein, SULFATE ION, ZINC ION | | Authors: | Stein, A.J, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a protein in the NADB-Rossmann Superfamily from Streptococcus agalactiae to 1.8A
To be Published
|
|
3LOF
 
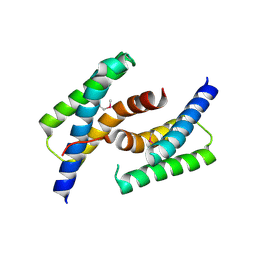 | | C-terminal domain of human heat shock 70kDa protein 1B. | | Descriptor: | Heat shock 70 kDa protein 1 | | Authors: | Osipiuk, J, Gu, M, Mihelic, M, Orton, K, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-03 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of C-terminal domain of human heat shock 70kDa protein 1B.
To be Published
|
|
3LP5
 
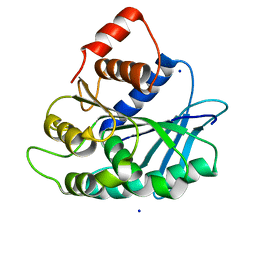 | |
3LAX
 
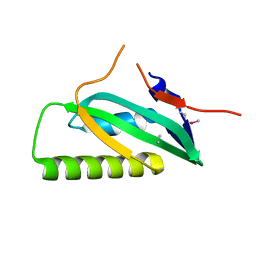 | |
3LQN
 
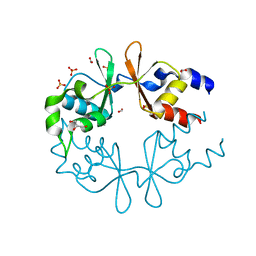 | | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor | | Descriptor: | CBS domain protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
3LR1
 
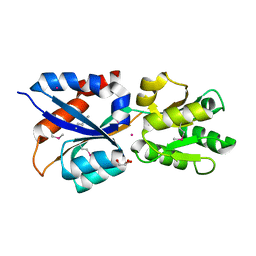 | | The crystal structure of the tungstate ABC transporter from Geobacter sulfurreducens | | Descriptor: | GLYCEROL, TUNGSTEN ION, Tungstate ABC transporter, ... | | Authors: | Zhang, R, Volkart, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the tungstate ABC transporter from Geobacter sulfurreducens
To be Published
|
|
3LFR
 
 | |
3LAE
 
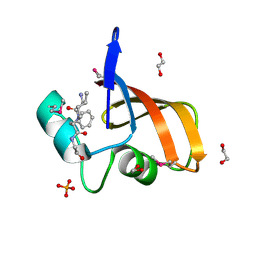 | | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, UPF0053 protein HI0107, ... | | Authors: | Tan, K, Li, H, Bargassa, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3LAO
 
 | |
3LGC
 
 | | Crystal Structure of Glutaredoxin 1 from Francisella tularensis | | Descriptor: | GLYCEROL, Glutaredoxin 1, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal Structure of Glutaredoxin 1 from Francisella tularensis
To be Published
|
|
3LJK
 
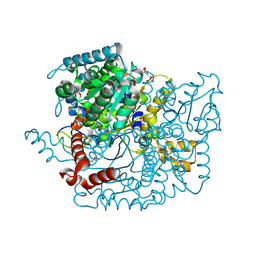 | | Glucose-6-phosphate isomerase from Francisella tularensis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | X-ray crystal structure of glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3LUP
 
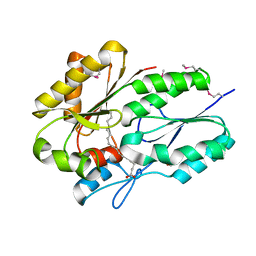 | | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae | | Descriptor: | 9-OCTADECENOIC ACID, DegV family protein, GLYCEROL | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae
To be Published
|
|
3LAZ
 
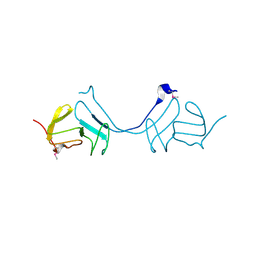 | |
3LOD
 
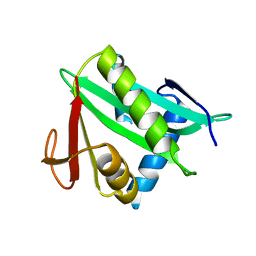 | |
3LVU
 
 | | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ABC transporter, ... | | Authors: | Chang, C, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi
To be Published
|
|
3LXY
 
 | | Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92 | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Nocek, B, Maltseva, N, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92
To be Published
|
|
