3DO8
 
 | |
3DOA
 
 | |
3DS8
 
 | |
3GOS
 
 | | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Yersinia pestis CO92 | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, MAGNESIUM ION | | Authors: | Zhang, R, Maltseva, N, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Yersinia pestis CO92
To be Published
|
|
3GVE
 
 | | Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Kim, Y, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Calcineurin-like Phosphoesterase from Bacillus subtilis
To be Published
|
|
3H2Z
 
 | |
3CP3
 
 | |
3D3S
 
 | | Crystal structure of L-2,4-diaminobutyric acid acetyltransferase from Bordetella parapertussis | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Kim, Y, Volkart, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of L-2,4-diaminobutyric acid acetyltransferase from Bordetella parapertussis.
To be Published
|
|
3CP0
 
 | | Crystal structure of the soluble domain of membrane protein implicated in regulation of membrane protease activity from Corynebacterium glutamicum | | Descriptor: | CHLORIDE ION, Membrane protein implicated in regulation of membrane protease activity, ZINC ION | | Authors: | Kim, Y, Tesar, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-30 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the soluble domain of membrane protein implicated in regulation of membrane protease activity from Corynebacterium glutamicum.
To be Published
|
|
3CRJ
 
 | |
3GYK
 
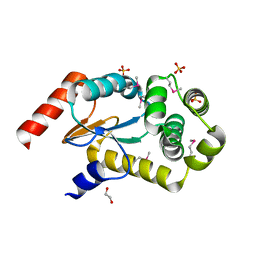 | | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3 | | Descriptor: | 1,2-ETHANEDIOL, 27kDa outer membrane protein, SULFATE ION | | Authors: | Fan, Y, Marshall, N, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3
To be Published
|
|
3GN3
 
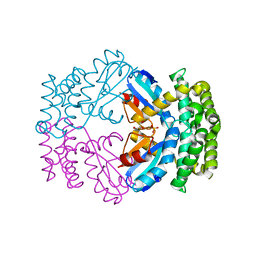 | | Crystal structure of a putative protein-disulfide isomerase from Pseudomonas syringae to 2.5A resolution. | | Descriptor: | GLYCEROL, SULFATE ION, putative protein-disulfide isomerase | | Authors: | Stein, A.J, Chhor, G, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-16 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative protein-disulfide isomerase from Pseudomonas syringae to 2.5A resolution.
To be Published
|
|
3EUS
 
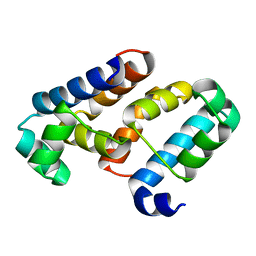 | |
3EEH
 
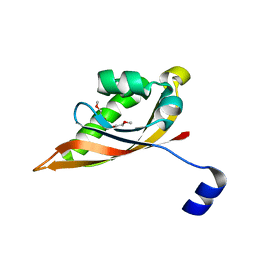 | |
3EFA
 
 | | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chang, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum
To be Published
|
|
3ERM
 
 | |
3EUP
 
 | | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii | | Descriptor: | SULFATE ION, Transcriptional regulator, TetR family | | Authors: | Zhang, R, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii
To be Published
|
|
3GVD
 
 | | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETIC ACID, CYSTEINE, ... | | Authors: | Kim, Y, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-30 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis
To be Published
|
|
3H1Q
 
 | | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ethanolamine utilization protein EutJ | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Kinney, J, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans
To be Published
|
|
3H0X
 
 | | Crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae | | Descriptor: | 78 kDa glucose-regulated protein homolog | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae.
To be Published
|
|
3GO9
 
 | | Predicted insulinase family protease from Yersinia pestis | | Descriptor: | DI(HYDROXYETHYL)ETHER, ZINC ION, insulinase family protease | | Authors: | Osipiuk, J, Maltseva, N, Onopriyenko, O, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structure of predicted insulinase family protease from Yersinia pestis.
To be Published
|
|
3H1S
 
 | | Crystal structure of superoxide dismutase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | FE (III) ION, GLYCEROL, Superoxide dismutase | | Authors: | Nocek, B, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of superoxide dismutase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3GX1
 
 | |
3GJY
 
 | |
3H07
 
 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Yersinia pestis CO92 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase | | Authors: | Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-08 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Yersinia pestis CO92
To be Published
|
|
