3FH2
 
 | |
3FIX
 
 | | Crystal structure of a putative n-acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYLTRANSFERASE | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3FX3
 
 | | Structure of a putative cAMP-binding regulatory protein from Silicibacter pomeroyi DSS-3 | | Descriptor: | Cyclic nucleotide-binding protein, GLYCEROL, PHOSPHATE ION | | Authors: | Cuff, M.E, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative cAMP-binding regulatory protein from Silicibacter pomeroyi DSS-3
TO BE PUBLISHED
|
|
3G0M
 
 | | Crystal structure of cysteine desulfuration protein SufE from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Cysteine desulfuration protein sufE, ... | | Authors: | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of cysteine desulfuration protein SufE from Salmonella typhimurium LT2
To be Published
|
|
3FZ4
 
 | | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative arsenate reductase, ... | | Authors: | Tan, K, Hatzos, C, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159.
To be Published
|
|
3FVV
 
 | |
3FWX
 
 | | The crystal structure of the peptide deformylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Zhang, R, Zhou, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the peptide deformylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3G0S
 
 | | Dihydrodipicolinate synthase from Salmonella typhimurium LT2 | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structure of dihydrodipicolinate synthase from Salmonella typhimurium LT2.
To be Published
|
|
3FTB
 
 | |
3EEV
 
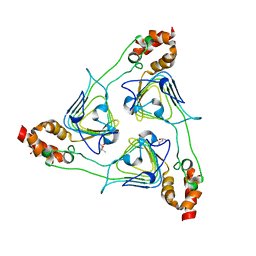 | | Crystal Structure of Chloramphenicol Acetyltransferase VCA0300 from Vibrio cholerae O1 biovar eltor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chloramphenicol acetyltransferase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Chloramphenicol Acetyltransferase VCA0300 from Vibrio cholerae O1 biovar eltor
To be Published
|
|
3EUR
 
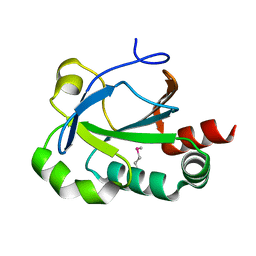 | |
3E58
 
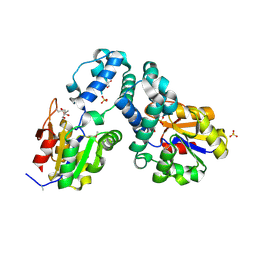 | | Crystal structure of putative beta-phosphoglucomutase from Streptococcus thermophilus | | Descriptor: | GLYCEROL, SULFATE ION, putative Beta-phosphoglucomutase | | Authors: | Chang, C, Tesar, C, Souvong, K, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-13 | | Release date: | 2008-08-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of putative beta-phosphoglucomutase from Streptococcus thermophilus
To be Published
|
|
3GAA
 
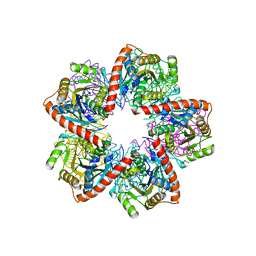 | |
3FQ6
 
 | |
3FZ5
 
 | | Crystal structure of possible 2-hydroxychromene-2-carboxylate isomerase from Rhodobacter sphaeroides | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Chang, C, Hatzos, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of possible 2-hydroxychromene-2-carboxylate isomerase from Rhodobacter sphaeroides
To be Published
|
|
3G1C
 
 | | The crystal structure of a TrpR like protein from Eubacterium eligens ATCC 27750 | | Descriptor: | MAGNESIUM ION, The TrpR like protein from Eubacterium eligens ATCC 27750 | | Authors: | Zhang, R, Hendricks, R, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a TrpR like protein from Eubacterium eligens ATCC 27750
To be Published
|
|
3FW2
 
 | | C-terminal domain of putative thiol-disulfide oxidoreductase from Bacteroides thetaiotaomicron. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, thiol-disulfide oxidoreductase | | Authors: | Osipiuk, J, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-16 | | Release date: | 2009-01-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray crystal structure of C-terminal domain of putative thiol-disulfide oxidoreductase from Bacteroides thetaiotaomicron.
To be Published
|
|
3FWW
 
 | | The crystal structure of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase from Yersinia pestis CO92 | | Descriptor: | Bifunctional protein glmU | | Authors: | Zhang, R, Gu, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase from Yersinia pestis CO92
To be Published
|
|
3D0K
 
 | | Crystal structure of the LpqC, poly(3-hydroxybutyrate) depolymerase from Bordetella parapertussis | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative poly(3-hydroxybutyrate) depolymerase LpqC, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the LpqC, Poly(3-hydroxybutyrate) Depolymerase from Bordetella parapertussis.
To be Published
|
|
3D1L
 
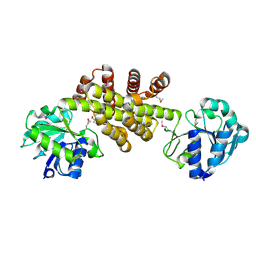 | | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis | | Descriptor: | 2-MERCAPTO-PROPION ALDEHYDE, CHLORIDE ION, Putative NADP oxidoreductase BF3122 | | Authors: | Chang, C, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis.
To be Published
|
|
3CNE
 
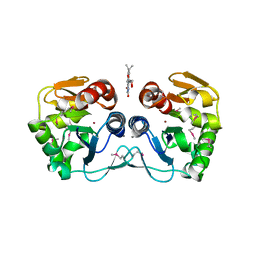 | | Crystal structure of the putative protease I from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Putative protease I, ... | | Authors: | Zhang, R, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the putative protease I from Bacteroides thetaiotaomicron.
To be Published
|
|
3D7L
 
 | |
3CJN
 
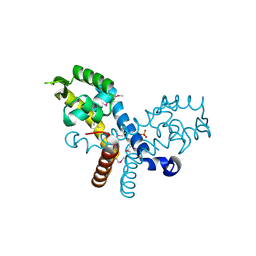 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, MarR family | | Authors: | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
3D6K
 
 | | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Tan, K, Zhang, R, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae
To be Published
|
|
3DNP
 
 | | Crystal structure of Stress response protein yhaX from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Stress response protein yhaX | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Stress response protein yhaX from Bacillus subtilis
To be Published
|
|
