5WHC
 
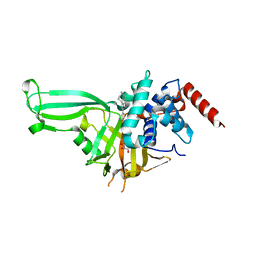 | | USP7 in complex with Cpd2 (4-(3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl)phenol) | | Descriptor: | 4-[3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl]phenol, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-07-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of Ubiquitin Specific Protease 7 (USP7) Using Integrated NMR and in Silico Techniques.
J. Med. Chem., 60, 2017
|
|
1OBM
 
 | |
6IVE
 
 | |
4PHW
 
 | |
4P1R
 
 | |
7B7V
 
 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
4TPP
 
 | | 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors | | Descriptor: | 1-[4-(3-{[1-(quinolin-2-yl)azetidin-3-yl]oxy}quinoxalin-2-yl)piperidin-1-yl]ethanone, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-06-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility.
Bioorg.Med.Chem., 22, 2014
|
|
4TPM
 
 | |
5KBX
 
 | |
6M3C
 
 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
6M3B
 
 | | hAPC-c25k23 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, ... | | Authors: | Wang, X, Li, L, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
7YHW
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.12.1 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
4N7M
 
 | |
4N7J
 
 | |
4NBN
 
 | |
4N5D
 
 | | Tailoring Small Molecules for an Allosteric Site on Procaspase-6: Cpd1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-amino-2,8-dimethylpyrido[2,3-d]pyrimidin-7(8H)-one, Caspase-6, ... | | Authors: | Murray, J.M, Steffek, M. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tailoring small molecules for an allosteric site on procaspase-6.
Chemmedchem, 9, 2014
|
|
4N6G
 
 | |
4NBK
 
 | |
4NBL
 
 | |
4P0N
 
 | | Crystal structure of PDE10a with a novel Imidazo[4,5-b]pyridine inhibitor | | Descriptor: | GLYCEROL, N-[cis-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, N-[trans-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazo[4,5-b]pyridines as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
Acs Med.Chem.Lett., 5, 2014
|
|
6IHB
 
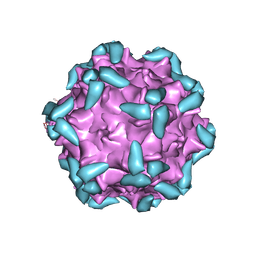 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
6J0W
 
 | | Crystal Structure of Yeast Rtt107 and Nse6 | | Descriptor: | Peptide from DNA repair protein KRE29, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0V
 
 | | Crystal Structure of Yeast Rtt107 | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0Y
 
 | | Crystal Structure of Yeast Rtt107 and Slx4 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0X
 
 | | Crystal Structure of Yeast Rtt107 and Mms22 | | Descriptor: | Peptide from E3 ubiquitin-protein ligase substrate receptor MMS22, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
