3A80
 
 | |
3A8C
 
 | |
3A7X
 
 | |
3A86
 
 | |
3B0U
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA fragment | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*(H2U)P*A)-3'), tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0P
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8H1K
 
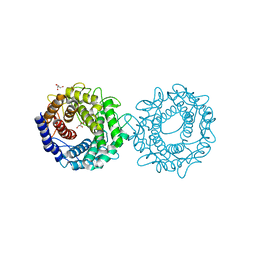 | | Crystal structure of glucose-2-epimerase from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, GLYCEROL, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1L
 
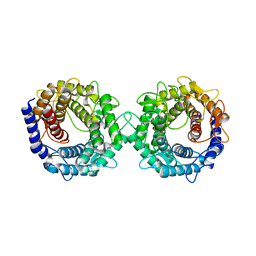 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1M
 
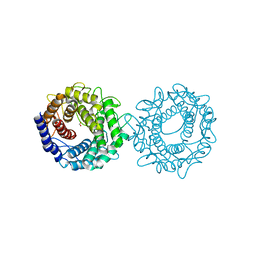 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
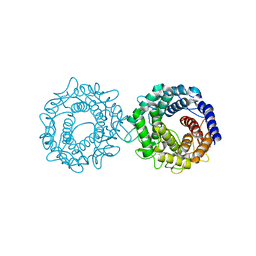 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8IN4
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
5Z3C
 
 | | Glycosidase E178A | | Descriptor: | GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3D
 
 | | Glycosidase F290Y | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3B
 
 | | Glycosidase Y48F | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3A
 
 | | Glycosidase Wild Type | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5ZTB
 
 | | Structure of Sulfurtransferase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Tanaka, Y, Chen, M, Narai, S, Yao, M. | | Deposit date: | 2018-05-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The [4Fe-4S] cluster of sulfurtransferase TtuA desulfurizes TtuB during tRNA modification in Thermus thermophilus.
Commun Biol, 3, 2020
|
|
5Z3E
 
 | | Glycosidase E335A | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3F
 
 | | Glycosidase E335A in complex with glucose | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5XFM
 
 | | Crystal structure of beta-arabinopyranosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Okuyama, M, Yao, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel glycoside hydrolase family 97 enzyme: Bifunctional beta-l-arabinopyranosidase/ alpha-galactosidase from Bacteroides thetaiotaomicron.
Biochimie, 142, 2017
|
|
5X3K
 
 | | Kfla1895 D451A mutant in complex with isomaltose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycoside hydrolase mutant in complex with product
To Be Published
|
|
8JH0
 
 | | Crystal structure of the light-driven sodium pump IaNaR | | Descriptor: | RETINAL, Xanthorhodopsin | | Authors: | Hashimoto, T, Kato, K, Tanaka, Y, Yao, M, Kikukawa, T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Multistep conformational changes leading to the gate opening of light-driven sodium pump rhodopsin.
J.Biol.Chem., 299, 2023
|
|
5X3J
 
 | | Kfla1895 D451A mutant in complex with cyclobis-(1->6)-alpha-nigerosyl | | Descriptor: | Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 31, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycoside hydrolase mutant in complex with substrate
To Be Published
|
|
