3NIT
 
 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O43
 
 | |
3O40
 
 | | Complex of a chimeric alpha/beta-peptide based on the gp41 CHR domain bound to gp41-5 | | Descriptor: | CHLORIDE ION, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2010-07-26 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
1DB3
 
 | | E.COLI GDP-MANNOSE 4,6-DEHYDRATASE | | Descriptor: | GDP-MANNOSE 4,6-DEHYDRATASE | | Authors: | Somoza, J.R, Menon, S, Somers, W.S, Sullivan, F.X. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme's catalytic mechanism and regulation by GDP-fucose.
Structure Fold.Des., 8, 2000
|
|
1HLA
 
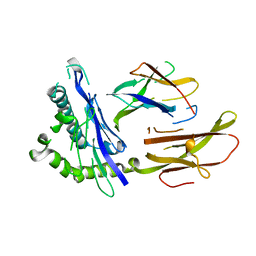 | | STRUCTURE OF THE HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN, HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2) (ALPHA CHAIN) | | Authors: | Bjorkman, P.J, Saper, M.A, Samraoui, B, Bennett, W.S, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1987-10-15 | | Release date: | 1988-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human class I histocompatibility antigen, HLA-A2.
Nature, 329, 1987
|
|
1DQE
 
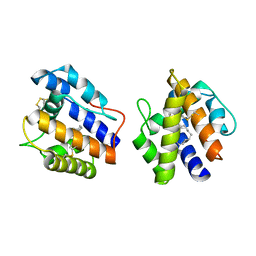 | | BOMBYX MORI PHEROMONE BINDING PROTEIN | | Descriptor: | HEXADECA-10,12-DIEN-1-OL, PHEROMONE-BINDING PROTEIN | | Authors: | Sandler, B.H, Nikonova, L, Leal, W.S, Clardy, J. | | Deposit date: | 2000-01-04 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex.
Chem.Biol., 7, 2000
|
|
1I7E
 
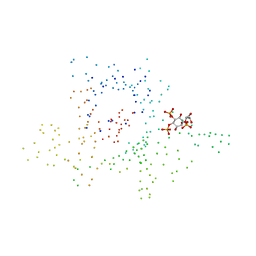 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
1F45
 
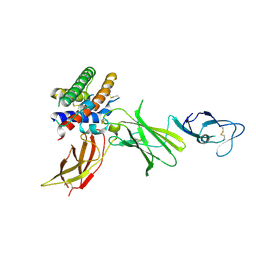 | | HUMAN INTERLEUKIN-12 | | Descriptor: | INTERLEUKIN-12 ALPHA CHAIN, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
1F46
 
 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1MX2
 
 | | Structure of F71N mutant of p18INK4c | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiery, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
4NIZ
 
 | | GCN4-p1 single Val9 to aminobutyric acid mutant | | Descriptor: | GLYCEROL, General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ2
 
 | | GCN4-p1 triple Val9, 23,30 to Ile mutant | | Descriptor: | GLYCEROL, General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
3G7A
 
 | | HIV gp41 six-helix bundle composed of a chimeric alpha+alpha/beta-peptide analogue of the CHR domain in complex with an NHR domain alpha-peptide | | Descriptor: | ACETYL GROUP, Chimeric alpha+alpha/beta-peptide analogue of the HIV gp41 CHR domain, Envelope glycoprotein gp160, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4OZB
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-ACPC24, beta-3-Lys28, beta-3-Lys31, beta-ACPC35 | | Descriptor: | GLYCEROL, Streptococcal Protein GB1 Backbone Modified Variant: beta-ACPC24, beta-3-Lys28, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
4OZC
 
 | | Backbone Modifications in the Protein GB1 Helix and Loops: beta-ACPC21, beta-ACPC24, beta-3-Lys28, beta-3-Lys31, beta-ACPC35, beta-ACPC40 | | Descriptor: | GLYCEROL, SULFATE ION, Streptococcal Protein GB1 Backbone Modified Variant: beta-ACPC21, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
3HF0
 
 | |
3HEZ
 
 | |
3HEY
 
 | | Cyclic residues in alpha/beta-peptide helix bundles: GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone with cyclic beta-residues at positions 1, 4, 10, 19 and 28 | | Descriptor: | ACETATE ION, alpha/beta-peptide based on the GCN4-pLI side chain sequence with an (alpha-alpha-beta) backbone and cyclic beta-residues at positions 1, 4, ... | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2009-05-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of beta-amino acid preorganization in a self-assembling alpha/beta-peptide: fundamental studies of foldameric helix bundles.
J.Am.Chem.Soc., 132, 2010
|
|
3HEW
 
 | |
3HEU
 
 | |
4NX9
 
 | | Crystal structure of Pseudomonas aeruginosa flagellin FliC | | Descriptor: | Flagellin | | Authors: | Song, W.S, Yoon, S.I. | | Deposit date: | 2013-12-09 | | Release date: | 2014-01-29 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament
Biochem.Biophys.Res.Commun., 444, 2014
|
|
4NJ0
 
 | | GCN4-p1 single Val9 to Ile mutant | | Descriptor: | General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ1
 
 | | GCN4-p1 double Val9, 23 to Ile mutant | | Descriptor: | General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4OZA
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Gln32, beta-3-Asp36 | | Descriptor: | ISOPROPYL ALCOHOL, Streptococcal Protein GB1 Backbone Modified Variant: beta-3-Ala24, beta-3-Lys28, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
4POO
 
 | | The crystal structure of Bacillus subtilis YtqB in complex with SAM | | Descriptor: | Putative RNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Park, S.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a putative SAM-dependent methyltransferase, YtqB, from Bacillus subtilis
Biochem.Biophys.Res.Commun., 446, 2014
|
|
