1X9T
 
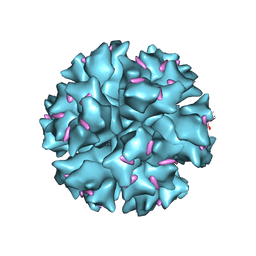 | | The crystal structure of human adenovirus 2 penton base in complex with an ad2 N-terminal fibre peptide | | Descriptor: | N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE, N-terminal peptide of Fiber protein, Penton protein | | Authors: | Zubieta, C, Schoehn, G, Chroboczek, J, Cusack, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the human adenovirus 2 penton
Mol.Cell, 17, 2005
|
|
1X9P
 
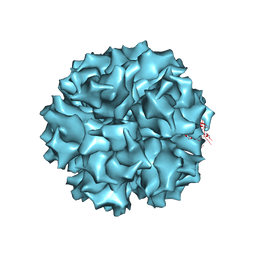 | | The crystal structure of human adenovirus 2 penton base | | Descriptor: | N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE, Penton protein, SULFATE ION | | Authors: | Zubieta, C, Schoehn, G, Chroboczek, J, Cusack, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of the human adenovirus 2 penton
Mol.Cell, 17, 2005
|
|
6FHI
 
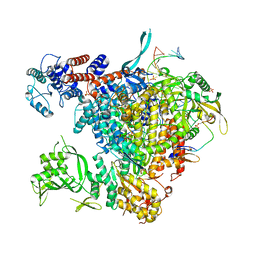 | |
5NG2
 
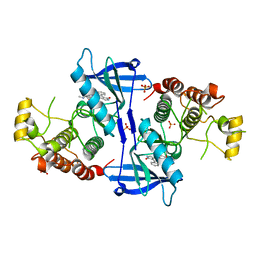 | | Structure of RIP2K(D146N) with bound Staurosporine | | Descriptor: | PHOSPHATE ION, Receptor-interacting serine/threonine-protein kinase 2, STAUROSPORINE | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
5NG0
 
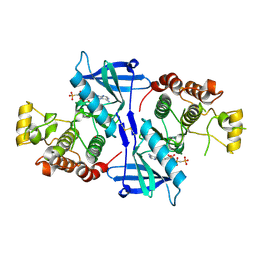 | | Structure of RIP2K(L294F) with bound AMPPCP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
5NG3
 
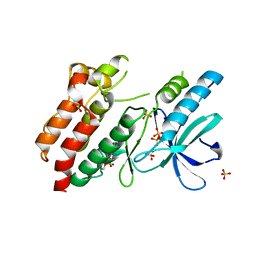 | | Structure of inactive kinase RIP2K(K47R) | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, SULFATE ION | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
1UXB
 
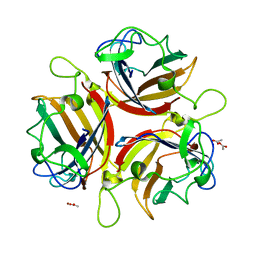 | | ADENOVIRUS AD19p FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
1UXE
 
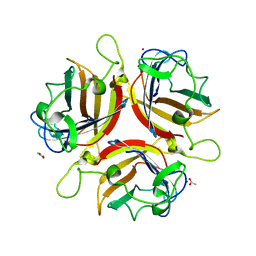 | | ADENOVIRUS AD37 FIBRE HEAD | | Descriptor: | ACETATE ION, FIBER PROTEIN, ZINC ION | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
2GMO
 
 | | NMR-structure of an independently folded C-terminal domain of influenza polymerase subunit PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Boudet, J, Tarendeau, F, Guilligay, D, Mas, P, Bougault, C.M, Cusack, S, Simorre, J.-P, Hart, D.J. | | Deposit date: | 2006-04-07 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1J1U
 
 | | Crystal structure of archaeal tyrosyl-tRNA synthetase complexed with tRNA(Tyr) and L-tyrosine | | Descriptor: | MAGNESIUM ION, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Kobayashi, T, Nureki, O, Ishitani, R, Tukalo, M, Cusack, S, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion
NAT.STRUCT.BIOL., 10, 2003
|
|
9HBX
 
 | |
9HBT
 
 | | TiLV-NP pentamer (pseudo-C5) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
9HBR
 
 | |
9HBV
 
 | |
9HBS
 
 | | TiLV-NP tetramer (pseudo-C2) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
9HBY
 
 | |
9HBU
 
 | |
9HBW
 
 | | TiLV-NP tetramer (pseudo-C4) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
9HBZ
 
 | | TiLV-NP hexamer (pseudo-C6) | | Descriptor: | 40-mer vRNA loop, Tilapia Lake Virus nucleoprotein (segment 4) | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2024-11-08 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the tilapia lake virus nucleoprotein bound to RNA.
Nucleic Acids Res., 53, 2025
|
|
7ORJ
 
 | | La Crosse virus polymerase at transcription capped RNA cleavage stage | | Descriptor: | 5' capped RNA, ADENOSINE-5'-TRIPHOSPHATE, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*CP*AP*AP*G)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORI
 
 | | La Crosse virus polymerase at replication late-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORO
 
 | | La Crosse virus polymerase at replication early-elongation stage | | Descriptor: | La Crosse virus polymerase, MAGNESIUM ION, RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORN
 
 | | La Crosse virus polymerase at replication initiation stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, La Crosse virus polymerase, MAGNESIUM ION, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORL
 
 | | La Crosse virus polymerase at transcription initiation stage | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
7ORM
 
 | | La Crosse virus polymerase at transcription early-elongation stage | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3'), RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*C)-3'), ... | | Authors: | Arragain, B, Durieux Trouilleton, Q, Baudin, F, Cusack, S, Schoehn, G, Malet, H. | | Deposit date: | 2021-06-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription.
Nat Commun, 13, 2022
|
|
