1J6R
 
 | |
4R1L
 
 | |
4R4K
 
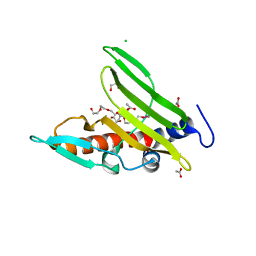 | | Crystal structure of a cystatin-like protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of a hypothetical protein (BACCAC_01506) from Bacteroides caccae ATCC 43185 at 1.69 A resolution
To be published
|
|
4R7S
 
 | |
1O5O
 
 | |
4RVN
 
 | |
4RU2
 
 | |
4RWV
 
 | | Crystal structure of PIP3 bound human nuclear receptor LRH-1 (Liver Receptor Homolog 1, NR5A2) in complex with a co-regulator DAX-1 (NR0B1) peptide at 1.86 A resolution | | Descriptor: | (2S)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology, Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Crystal structure of a Homo sapiens hepatocytic transcription factor hB1F-2 (B1F2) in complex with nuclear receptor subfamily 0 group B member 1 (NR0B1, residues 140-154) from human at 1.86 A resolution
To be published
|
|
1J5P
 
 | |
1J5X
 
 | |
4ORL
 
 | |
4GHB
 
 | |
4OBI
 
 | |
4OGZ
 
 | |
4G2A
 
 | |
1O54
 
 | |
4OPM
 
 | |
4OBM
 
 | |
4PWU
 
 | |
4HW6
 
 | |
4Q5T
 
 | |
4Q69
 
 | |
4IAJ
 
 | |
4PW1
 
 | |
4HSP
 
 | |
