8PP8
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with L-scyllo-inositol 1,2,4-trisphosphate/AMP-PNP/Mn | | Descriptor: | Inositol-trisphosphate 3-kinase A, L-scyllo-inositol 1,2,4-trisphosphate, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPA
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with D-myo-inositol 1,4,6-trisphosphate/AMP-PNP/Mn | | Descriptor: | D-myo-inositol 1,4,6-trisphosphate, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPD
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with DL-6-deoxy-6-hydroxy-methyl-scyllo-inositol 1,2,4-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DL-6-deoxy-6-hydroxy-methyl-scyllo-inositol 1,2,4-trisphosphate, Inositol-trisphosphate 3-kinase A, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPG
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosyl 1,3,4,6-tetrakisphosphate/ADP/Mn after reaction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPB
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with D-3-deoxy-myo-inositol 1,4,6-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-3-deoxy-myo-inositol 1,4,6-trisphosphate, Inositol-trisphosphate 3-kinase A, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPC
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with L-chiro-inositol 2,3,5-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-trisphosphate 3-kinase A, L-chiro-inositol 2,3,5-trisphosphate, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPF
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosyl 1,3,4-trisphosphate/AMP-PNP/Mg | | Descriptor: | Inositol-trisphosphate 3-kinase A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPE
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with DL-6-deoxy-6-phosphoryloxymethyl-scyllo-inositol 1,2,4-trisphosphate/ADP/Mn | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DL-6-deoxy-6-phosphoryloxymethyl-scyllo-inositol 1,2,4-trisphosphate, GLYCEROL, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
8PPI
 
 | | Human inositol 1,4,5-trisphosphate 3-kinase A (IP3K) catalytic domain in complex with beta-D-glucopyranosylmethanol 3,4,1'-trisphosphate/ATP/Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-trisphosphate 3-kinase A, MANGANESE (II) ION, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity.
Nat Commun, 15, 2024
|
|
5HIZ
 
 | | The structure of PEDV NSP9 | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
5HIY
 
 | | Crystal structure of PEDV NSP9 Mutant-C59A | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
3OWK
 
 | | Human CK2 catalytic domain in complex with a benzopyridoindole derivative inhibitor | | Descriptor: | 7-chloro-10-methyl-11H-benzo[g]pyrido[4,3-b]indol-3-ol, CSNK2A1 protein, SULFATE ION | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2.
Cancer Res., 70, 2010
|
|
3OWJ
 
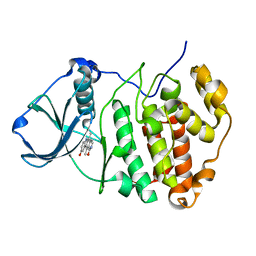 | | Human CK2 catalytic domain in complex with a pyridocarbazole derivative inhibitor | | Descriptor: | 9-hydroxy-5,11-dimethyl-4,6-dihydro-1H-pyrido[4,3-b]carbazol-1-one, CSNK2A1 protein | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2.
Cancer Res., 70, 2010
|
|
3OWL
 
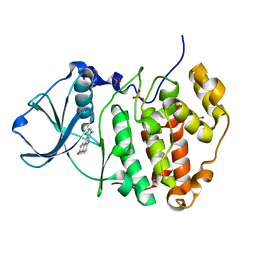 | | Human CK2 catalytic domain in complex with a benzopyridoindole derivative inhibitor | | Descriptor: | 11-chloro-8-methyl-7H-benzo[e]pyrido[4,3-b]indol-3-ol, CSNK2A1 protein, SULFATE ION | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2.
Cancer Res., 70, 2010
|
|
5YM8
 
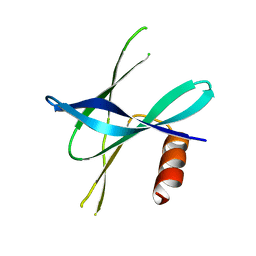 | |
5YM6
 
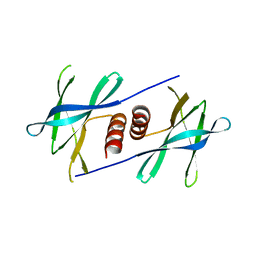 | |
6IVD
 
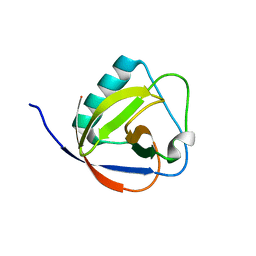 | | TGEV nsp1 mutant - 91-95sg | | Descriptor: | nsp1 mutant protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
6IVC
 
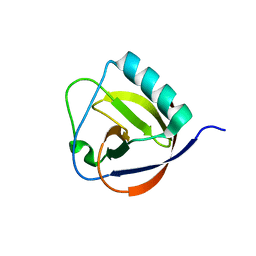 | | The full length of TGEV nsp1 | | Descriptor: | nsp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
7VGT
 
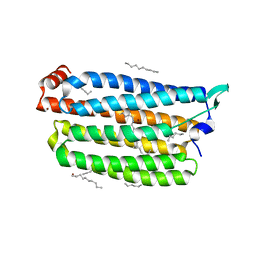 | | Time-resolved serial femtosecond crystallography structure of light-driven chloride ion-pumping rhodopsin, NM-R3: resting state structure with bromide ion | | Descriptor: | BROMIDE ION, Chloride pumping rhodopsin, DECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGV
 
 | | Anion free form of light-driven chloride ion-pumping rhodopsin, NM-R3, structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, HEXADECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGU
 
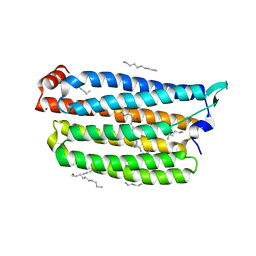 | | Time-resolved serial femtosecond crystallography structure of light-driven chloride ion-pumping rhodopsin, NM-R3 : structure obtained 1 msec after photoexcitation with bromide ion | | Descriptor: | BROMIDE ION, Chloride pumping rhodopsin, DECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
