5M8W
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 4-chlorophenol | | Descriptor: | 4-chlorophenol, BENZAMIDINE, GLYCEROL, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-10-30 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
5MC0
 
 | | Crystal Structure of delTyr231 mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Wilk, P, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
5MC2
 
 | | Crystal Structure of Gly278Asp mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
5MTT
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MalE1, ... | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5MTU
 
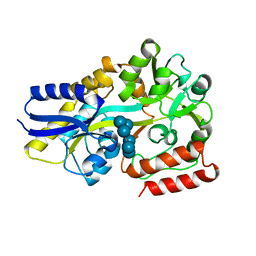 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
2X9A
 
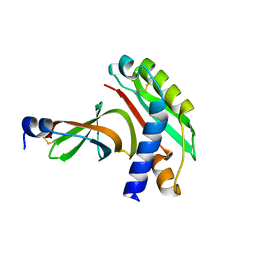 | | crystal structure of g3p from phage IF1 in complex with its coreceptor, the C-terminal domain of TolA | | Descriptor: | ATTACHMENT PROTEIN G3P, MEMBRANE SPANNING PROTEIN, REQUIRED FOR OUTER MEMBRANE INTEGRITY | | Authors: | Lorenz, S.H, Jakob, R.P, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
2X9B
 
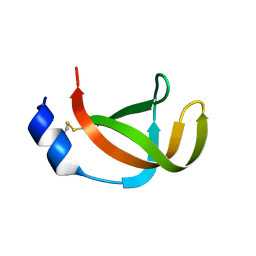 | | The filamentous phages fd and IF1 use different infection mechanisms | | Descriptor: | ATTACHMENT PROTEIN G3P | | Authors: | Lorenz, S.H, Jakob, R.P, Weininger, U, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
5OBP
 
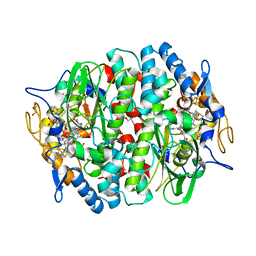 | | PCE reductive dehalogenase from S. multivorans with 6-hydroxybenzimidazolyl norcobamide cofactor | | Descriptor: | 6-hydroxybenzimidazolyl-norcobamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Keller, S, Kunze, C, Bommer, M, Paetz, C, Menezes, R.C, Svatos, A, Dobbek, H, Schubert, T. | | Deposit date: | 2017-06-28 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Selective Utilization of Benzimidazolyl-Norcobamides as Cofactors by the Tetrachloroethene Reductive Dehalogenase of Sulfurospirillum multivorans.
J. Bacteriol., 200, 2018
|
|
5OBI
 
 | | PCE reductive dehalogenase from S. multivorans with 5-METHOXYBENZIMIDAZOLYL-NORCOBAMIDE cofactor | | Descriptor: | 5-Methoxybenzimidazolyl-norcobamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Keller, S, Kunze, C, Bommer, M, Paetz, C, Menezes, R.C, Svatos, A, Dobbek, H, Schubert, T. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Selective Utilization of Benzimidazolyl-Norcobamides as Cofactors by the Tetrachloroethene Reductive Dehalogenase of Sulfurospirillum multivorans.
J. Bacteriol., 200, 2018
|
|
2YIV
 
 | | NI,FE-CODH with n-butylisocyanate state | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (III) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | N-Butyl Isocyanide Oxidation at the [Nife4S4Oh(X)] Cluster of Co Dehydrogenase.
J.Biol.Inorg.Chem., 17, 2012
|
|
2YCK
 
 | | methyltransferase bound with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 5-METHYLTETRAHYDROFOLATE CORRINOID/IRON SULFUR PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Goetzl, S, Jeoung, J.-H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
2YCJ
 
 | | methyltransferase bound with methyltetrahydrofolate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-METHYLTETRAHYDROFOLATE CORRINOID/IRON SULFUR PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Goetzl, S, Jeoung, J.-H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
2YCI
 
 | | methyltransferase native | | Descriptor: | 5-METHYLTETRAHYDROFOLATE CORRINOID/IRON SULFUR PROTEIN METHYLTRANSFERASE, SULFATE ION | | Authors: | Goetzl, S, Jeoung, J.H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
2YCL
 
 | | complete structure of the corrinoid,iron-sulfur protein including the N-terminal domain with a 4Fe-4S cluster | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE CORRINOID/IRON-SULFUR PROTEIN, GAMMA SUBUNIT, CO DEHYDROGENASE/ACETYL-COA SYNTHASE, ... | | Authors: | Goetzl, S, Jeoung, J.H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
3B51
 
 | | Ni,Fe-CODH-600 mV state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2007-10-25 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase.
Science, 318, 2007
|
|
3B53
 
 | | Ni,Fe-CODH-320 mV state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2007-10-25 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase.
Science, 318, 2007
|
|
3B52
 
 | | Ni,Fe-CODH-600 mV state + CO2 | | Descriptor: | CARBON DIOXIDE, Carbon monoxide dehydrogenase 2, FE (II) ION, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2007-10-25 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase.
Science, 318, 2007
|
|
6H2P
 
 | | Crystal Structure of Arg184Gln mutant of Human Prolidase with Mn ions and Cacodylate ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Piwowarczyk, R, Weiss, M.S. | | Deposit date: | 2018-07-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
6H2Q
 
 | | Crystal Structure of Arg184Gln mutant of Human Prolidase with Mn ions and LeuPro ligand | | Descriptor: | GLYCEROL, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Piwowarczyk, R, Weiss, M.S. | | Deposit date: | 2018-07-14 | | Release date: | 2018-08-15 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
7PPV
 
 | |
7PPT
 
 | |
7PPU
 
 | |
6ST5
 
 | | crystal structure of LicM2 | | Descriptor: | GLYCEROL, LicM2, MAGNESIUM ION, ... | | Authors: | Gonsior, M, Mainz, A, Hugelland, M, Kuthning, A, Tietzmann, M, Dobbek, H, Martins, B.M, Sussmuth, R. | | Deposit date: | 2019-09-10 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | crystal structure of LicM2
To Be Published
|
|
5NSF
 
 | | Structure of AzuAla | | Descriptor: | (2~{S})-2-azanyl-3-(2,6-dihydroazulen-1-yl)propanoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Martins, B.M. | | Deposit date: | 2017-04-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Site-Resolved Observation of Vibrational Energy Transfer Using a Genetically Encoded Ultrafast Heater.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
2YMV
 
 | | Structure of Reduced M Smegmatis 5246, a homologue of M.Tuberculosis Acg | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, ACG NITROREDUCTASE, ... | | Authors: | Chauviac, F.-X, Bommer, M, Dobbek, H, Keep, N.H. | | Deposit date: | 2012-10-10 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Reduced Msacg, a Putative Nitroreductase from Mycobacterium Smegmatis and a Close Homologue of Mycobacterium Tuberculosis Acg.
J.Biol.Chem., 287, 2012
|
|
