8TXF
 
 | | AvrB bound with RIN4 C-NOI motif | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avirulence protein B, ... | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
8TWJ
 
 | | AvrB_R266A bound with UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avirulence protein B, ... | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
8TWO
 
 | | AvrB bound with UDP and RIN4_T166-Rha | | Descriptor: | Avirulence protein B, RPM1-interacting protein 4, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
1MMD
 
 | | TRUNCATED HEAD OF MYOSIN FROM DICTYOSTELIUM DISCOIDEUM COMPLEXED WITH MGADP-BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Holden, H.M, Rayment, I. | | Deposit date: | 1995-03-21 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4-.
Biochemistry, 34, 1995
|
|
4DI3
 
 | |
4DI4
 
 | | Crystal structure of a 3:1 complex of Treponema pallidum TatP(T) (Tp0957) bound to TatT (Tp0956) | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, TatP(T) (Tp0957), ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-01-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.714 Å) | | Cite: | Structural and Thermodynamic Characterization of the Interaction between Two Periplasmic Treponema pallidum Lipoproteins that are Components of a TPR-Protein-Associated TRAP Transporter (TPAT).
J.Mol.Biol., 420, 2012
|
|
1ECG
 
 | | DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE | | Descriptor: | 5-OXO-L-NORLEUCINE, GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE AMIDOTRANSFERASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Krahn, J.M. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site.
J.Biol.Chem., 271, 1996
|
|
3C17
 
 | |
2N1T
 
 | | Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution | | Descriptor: | Synaptosomal-associated protein 25, Synaptotagmin-1, Syntaxin-1A, ... | | Authors: | Brewer, K, Bacaj, T, Cavalli, A, Camilloni, C, Swarbrick, J, Liu, J, Zhou, A, Zhou, P, Barlow, N, Xu, J, Seven, A, Prinslow, E, Voleti, R, Haussinger, D, Bonvin, A, Tomchick, D, Vendruscolo, M, Graham, B, Sudhof, T, Rizo, J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8USO
 
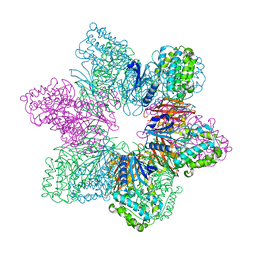 | | Full-length human CaMKII delta holoenzyme | | Descriptor: | MALONATE ION, calcium/calmodulin-dependent protein kinase | | Authors: | Ozden, C, Abromson, N.L, Tomchick, D.R, Stratton, M.M, Garman, S.C. | | Deposit date: | 2023-10-28 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human CaMKII delta holoenzyme
To be published
|
|
2ZAL
 
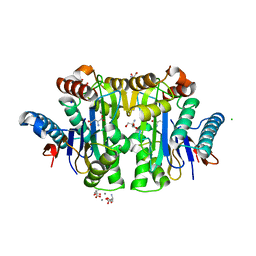 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
2ZAK
 
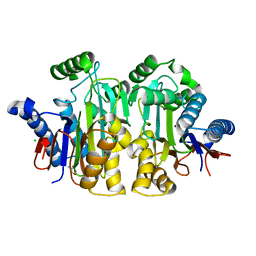 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7KH2
 
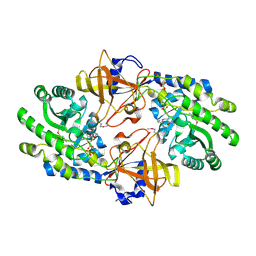 | | Structure of N-citrylornithine decarboxylase bound with PLP | | Descriptor: | GLYCEROL, N-citrylornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M, Michael, A. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-16 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Alternative pathways utilize or circumvent putrescine for biosynthesis of putrescine-containing rhizoferrin.
J.Biol.Chem., 296, 2020
|
|
7L0K
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM784 (3-(1-(3-methyl-4-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrrole-2-carboxamido)ethyl)-1H-pyrazole-5-carboxamide) | | Descriptor: | 3-{(1R)-1-[(3-methyl-4-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-1H-pyrrole-2-carbonyl)amino]ethyl}-1H-pyrazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM778 (3-methyl-N-(1-(5-methyl-1H-pyrazol-3-yl)ethyl)-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1H-pyrazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM589 (ethyl 3-methyl-4-((4-(trifluoromethyl)benzo[d]oxazol-7-yl)methyl)-1H-pyrrole-2-carboxylate) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZ4
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM705 (N-(1-(1H-1,2,4-triazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(1H-1,2,4-triazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYY
 
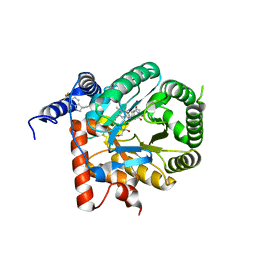 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM697 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(6-(trifluoromethyl)-1H-indol-3-yl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-[6-(trifluoromethyl)-1H-indol-3-yl]-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7L01
 
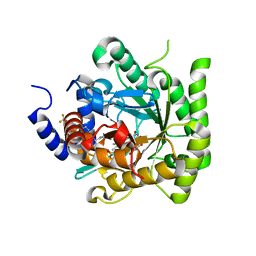 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM782 (N-(1-(5-cyano-1H-pyrazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYV
 
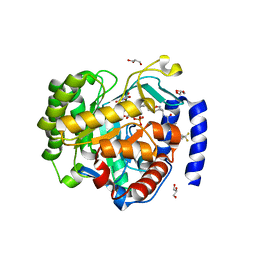 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM634 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(4-(trifluoromethyl)benzyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-{[4-(trifluoromethyl)phenyl]methyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
2GEZ
 
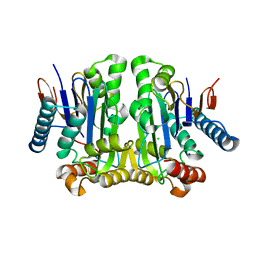 | | Crystal structure of potassium-independent plant asparaginase | | Descriptor: | CHLORIDE ION, L-asparaginase alpha subunit, L-asparaginase beta subunit, ... | | Authors: | Michalska, K, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant asparaginase.
J.Mol.Biol., 360, 2006
|
|
2UXX
 
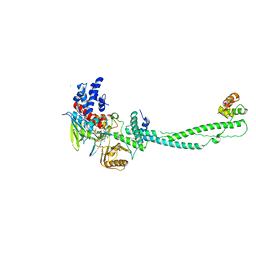 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
6AR0
 
 | | Structure of human SLMAP FHA domain | | Descriptor: | Sarcolemmal membrane-associated protein | | Authors: | Ni, L, Luo, X. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK.
Elife, 6, 2017
|
|
6AR2
 
 | |
1I84
 
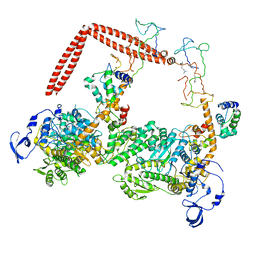 | | CRYO-EM STRUCTURE OF THE HEAVY MEROMYOSIN SUBFRAGMENT OF CHICKEN GIZZARD SMOOTH MUSCLE MYOSIN WITH REGULATORY LIGHT CHAIN IN THE DEPHOSPHORYLATED STATE. ONLY C ALPHAS PROVIDED FOR REGULATORY LIGHT CHAIN. ONLY BACKBONE ATOMS PROVIDED FOR S2 FRAGMENT. | | Descriptor: | SMOOTH MUSCLE MYOSIN ESSENTIAL LIGHT CHAIN, SMOOTH MUSCLE MYOSIN HEAVY CHAIN, SMOOTH MUSCLE MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Wendt, T, Taylor, D, Trybus, K.M, Taylor, K. | | Deposit date: | 2001-03-12 | | Release date: | 2001-03-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (20 Å) | | Cite: | Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
