2NQJ
 
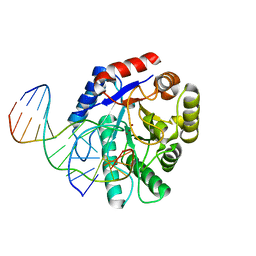 | | Crystal structure of Escherichia coli endonuclease IV (Endo IV) E261Q mutant bound to damaged DNA | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', Endonuclease 4, ... | | Authors: | Garcin-Hosfield, E.D, Hosfield, D.J, Tainer, J.A. | | Deposit date: | 2006-10-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | DNA apurinic-apyrimidinic site binding and excision by endonuclease IV.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2NQ9
 
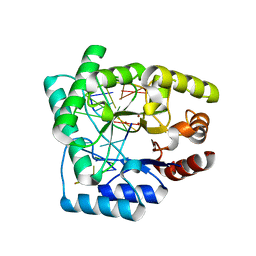 | | High resolution crystal structure of Escherichia coli endonuclease IV (Endo IV) Y72A mutant bound to damaged DNA | | Descriptor: | 5'-D(*AP*TP*AP*TP*CP*T)-3', 5'-D(*AP*TP*CP*TP*GP*AP*AP*GP*TP*AP*T)-3', 5'-D(P*(3DR)P*AP*GP*AP*T)-3', ... | | Authors: | Garcin-Hosfield, E.D, Hosfield, D.J, Tainer, J.A. | | Deposit date: | 2006-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA apurinic-apyrimidinic site binding and excision by endonuclease IV.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3QKS
 
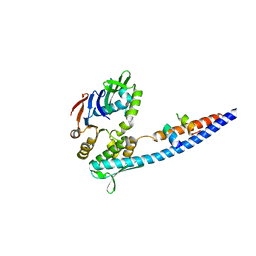 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1M9J
 
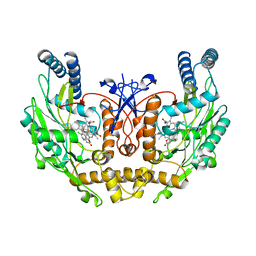 | | human endothelial nitric oxide synthase with chlorzoxazone bound | | Descriptor: | CHLORZOXAZONE, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
2R27
 
 | | Constitutively zinc-deficient mutant of human superoxide dismutase (SOD), C6A, H80S, H83S, C111S | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Roberts, B.R, Getzoff, E.D, Karplus, P.A, Beckman, J.S, Tainer, J.A. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS.
J.Mol.Biol., 373, 2007
|
|
2QT2
 
 | | Cyclized-Dehydrated Intermediate of GFP Variant Q183E in Chromophore Maturation | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-07-31 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetically Isolated Reaction Intermediates Provide Structural Characterization of the Green Fluorescence Protein Fluorophore Biosynthesis Pathway
To be Published
|
|
1M8I
 
 | | inducible nitric oxide synthase with 5-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M8E
 
 | | inducible nitric oxide synthase with 7-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M9M
 
 | | human endothelial nitric oxide synthase with 6-nitroindazole bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-NITROINDAZOLE, ISOPROPYL ALCOHOL, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
3QKT
 
 | | Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP | | Descriptor: | DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1M8H
 
 | | inducible nitric oxide synthase with 6-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
1M9K
 
 | | Human Endothelial Nitric Oxide Synthase with 7-Nitroindazole Bound | | Descriptor: | 7-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-29 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
2QRF
 
 | | Green Fluorescent Protein: Cyclized-only Intermediate of Chromophore Maturation in the Q183E variant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-07-28 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Chromophore - Cyclized Intermediate
To be Published
|
|
1M9A
 
 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with S-hexylglutathione | | Descriptor: | Glutathione S-Transferase 26 kDa, S-HEXYLGLUTATHIONE | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
2NQH
 
 | |
2QZ0
 
 | | Mature Q183E variant of Green Fluorescent Protein Chromophore | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-08-15 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetically Isolated Reaction Intermediates Provide Structural Characterization of the GFP Fluorophore Biosynthesis Pathway
To be Published
|
|
1DF1
 
 | | MURINE INOSOXY DIMER WITH ISOTHIOUREA BOUND IN THE ACTIVE SITE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Rosenfeld, R.J, Arvai, A.S, Ghosh, D.K, Ghosh, S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-16 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
EMBO J., 18, 1999
|
|
1DKT
 
 | | CKSHS1: HUMAN CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1 COMPLEX WITH METAVANADATE | | Descriptor: | CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1, META VANADATE | | Authors: | Bourne, Y, Arvai, A.S, Tainer, J.A. | | Deposit date: | 1995-11-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain.
J.Mol.Biol., 249, 1995
|
|
1DKS
 
 | | CKSHS1: HUMAN CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1 IN COMPLEX WITH PHOSPHATE | | Descriptor: | CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1, PHOSPHATE ION | | Authors: | Bourne, Y, Arvai, A.S, Tainer, J.A. | | Deposit date: | 1995-11-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain.
J.Mol.Biol., 249, 1995
|
|
1EM1
 
 | | X-RAY CRYSTAL STRUCTURE FOR HUMAN MANGANESE SUPEROXIDE DISMUTASE, Q143A | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE, SULFATE ION | | Authors: | Leveque, V, Stroupe, M.E, Lepock, J.R, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2000-03-14 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Multiple replacements of glutamine 143 in human manganese superoxide dismutase: effects on structure, stability, and catalysis.
Biochemistry, 39, 2000
|
|
1EMJ
 
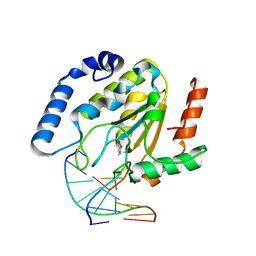 | | URACIL-DNA GLYCOSYLASE BOUND TO DNA CONTAINING A 4'-THIO-2'DEOXYURIDINE ANALOG PRODUCT | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(ASU)P*AP*TP*CP*TP*T)-3'), URACIL, ... | | Authors: | Parikh, S.S, Walcher, G, Jones, G.D, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4B22
 
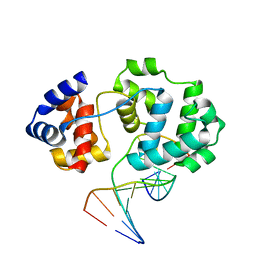 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*GP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP*(3DR)P*CP*AP*TP*CP*GP)-3', MAG2, ... | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
4B21
 
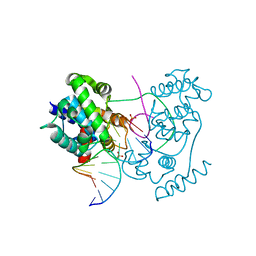 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*GP*AP*TP*CP*GP)-3', PHOSPHATE ION, ... | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
4EEU
 
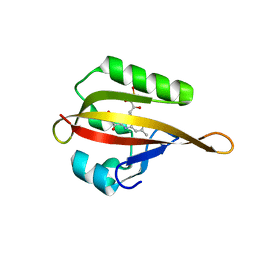 | | Crystal structure of phiLOV2.1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4068 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4ENJ
 
 | |
