1AI7
 
 | | PENICILLIN ACYLASE COMPLEXED WITH PHENOL | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, PHENOL | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1AJN
 
 | | PENICILLIN ACYLASE COMPLEXED WITH P-NITROPHENYLACETIC ACID | | Descriptor: | 2-(4-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJP
 
 | |
1AJQ
 
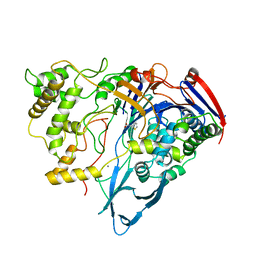 | | PENICILLIN ACYLASE COMPLEXED WITH THIOPHENEACETIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, THIOPHENEACETIC ACID | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1BNI
 
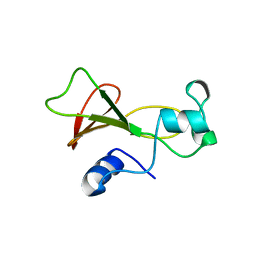 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1AI6
 
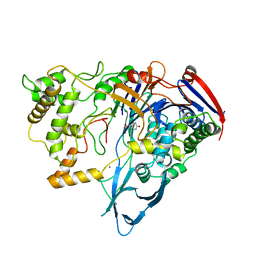 | | PENICILLIN ACYLASE WITH P-HYDROXYPHENYLACETIC ACID | | Descriptor: | 4-HYDROXYPHENYLACETATE, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI4
 
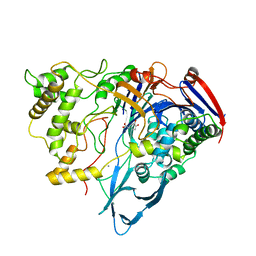 | |
1B9J
 
 | |
1BAO
 
 | |
1AQZ
 
 | |
1BGS
 
 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
1BNS
 
 | | STRUCTURAL STUDIES OF BARNASE MUTANTS | | Descriptor: | BARNASE | | Authors: | Chen, Y.W. | | Deposit date: | 1994-04-11 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of buried hydrogen bonds to protein stability. The crystal structures of two barnase mutants.
J.Mol.Biol., 234, 1993
|
|
1BRS
 
 | |
1BSE
 
 | |
1BSD
 
 | |
1BRG
 
 | |
1BSB
 
 | |
1B2Z
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BSA
 
 | |
1B20
 
 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B3S
 
 | |
1B2S
 
 | |
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BAN
 
 | |
