8IKM
 
 | | Trans complex of phospho parkin | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8IKT
 
 | | Ternary trans-complex of phospho-parkin with cis ACT and pUb | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-AMINOPROPANE, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8JWV
 
 | | Untethered R0RBR | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8IK6
 
 | | pUbl depleted Parkin complex with pUbiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, Ubiquitin, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
6IDN
 
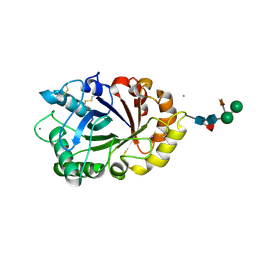 | | Crystal structure of ICChI chitinase from ipomoea carnea | | Descriptor: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | Authors: | Kumar, S, Kumar, A, Patel, A.K. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
7W9A
 
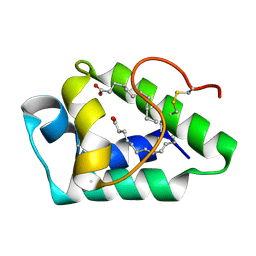 | |
7XQ7
 
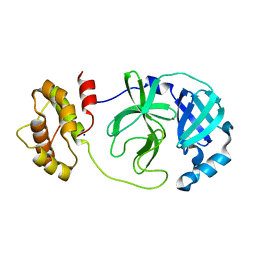 | | The complex structure of WT-Mpro | | Descriptor: | 3C-like proteinase nsp5, SODIUM ION | | Authors: | Sahoo, P, Lenka, D.R, Kumar, A. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7XQ6
 
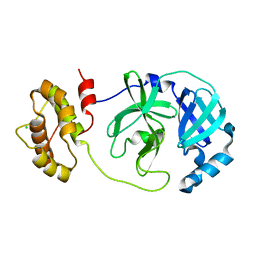 | | The complex structure of mutant Mpro with inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Sahoo, P, Lenka, D.R, Kumar, A. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7W9G
 
 | | Complex structure of Mpro with ebselen-derivative inhibitor | | Descriptor: | 3C-like proteinase nsp5, SELENIUM ATOM | | Authors: | Sahoo, P, Kumar, A. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
6KP1
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-methionine amino acid | | Descriptor: | METHIONINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KP0
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-arginine | | Descriptor: | ARGININE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KOZ
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323 mutant bound to L-Leucine amino acid | | Descriptor: | LEUCINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KOY
 
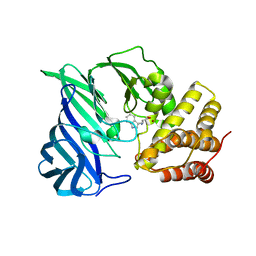 | | Crystal structure of two domain M1 Zinc metallopeptidase E323A mutant bound to L-tryptophan amino acid | | Descriptor: | TRYPTOPHAN, ZINC ION, Zinc metalloprotease | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
8ZN1
 
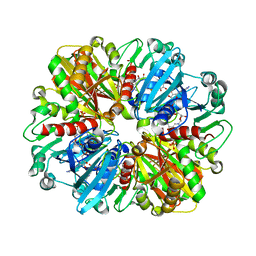 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
8ZN4
 
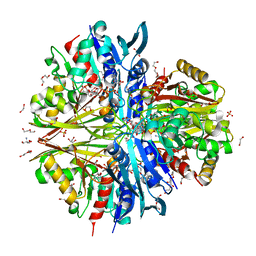 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
8ZOZ
 
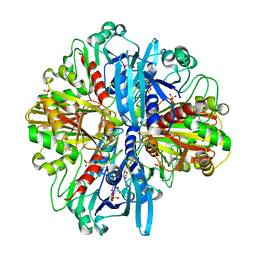 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
8WT1
 
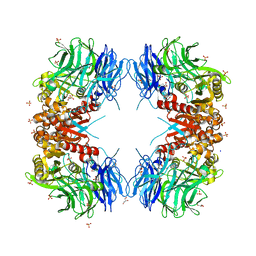 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
8HY5
 
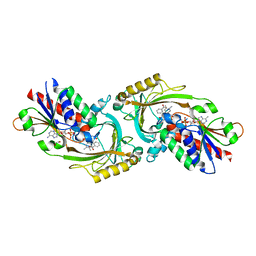 | | Structure of D-amino acid oxidase mutant R38H | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Khan, S, Upadhyay, S, Dave, U, Kumar, A, Gomes, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into ALS patient derived mutations in D-amino acid oxidase.
Int.J.Biol.Macromol., 256, 2023
|
|
7DF6
 
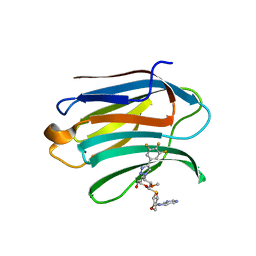 | | Mouse Galectin-3 CRD in complex with novel tetrahydropyran-based thiodisaccharide mimic inhibitor | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-5-methoxy-6-[(3R,4R,5S)-4-oxidanyl-5-(4-pyrimidin-5-yl-1,2,3-triazol-1-yl)oxan-3-yl]sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-ol, Galectin-3 | | Authors: | Ghosh, K, Kumar, A. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, Structure-Activity Relationships, and In Vivo Evaluation of Novel Tetrahydropyran-Based Thiodisaccharide Mimics as Galectin-3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7DF5
 
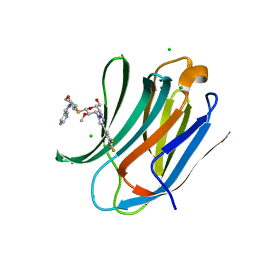 | | Human Galectin-3 CRD in complex with novel tetrahydropyran-based thiodisaccharide mimic inhibitor | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-5-methoxy-6-[(3R,4R,5S)-4-oxidanyl-5-(4-pyrimidin-5-yl-1,2,3-triazol-1-yl)oxan-3-yl]sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-ol, CHLORIDE ION, Galectin-3, ... | | Authors: | Ghosh, K, Kumar, A. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Synthesis, Structure-Activity Relationships, and In Vivo Evaluation of Novel Tetrahydropyran-Based Thiodisaccharide Mimics as Galectin-3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
3P20
 
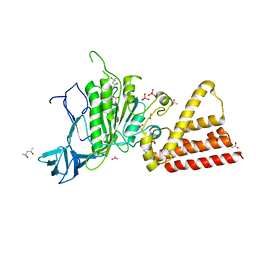 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-10-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
8TU9
 
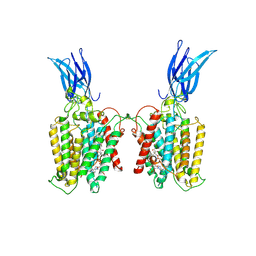 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Elife, 13, 2024
|
|
7B7F
 
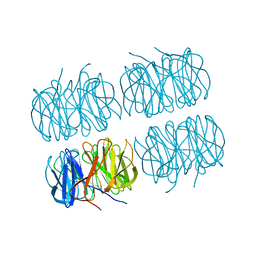 | | Room temperature X-ray structure of H/D-exchanged PLL lectin in complex with L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7B7C
 
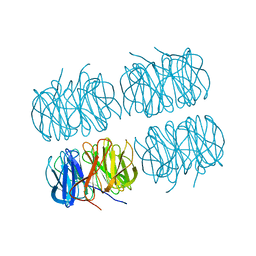 | | Room temperature X-ray structure of perdeuterated PLL lectin in complex with L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7B7E
 
 | | Room temperature X-ray structure of perdeuterated PLL lectin | | Descriptor: | PLL lectin | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
