2ZOF
 
 | | Crystal structure of mouse carnosinase CN2 complexed with MN and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Cytosolic non-specific dipeptidase, MANGANESE (II) ION | | Authors: | Unno, H, Yamashita, T, Okumura, N, Kusunoki, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate recognition and hydrolysis by mouse carnosinase CN2.
J.Biol.Chem., 283, 2008
|
|
3A7L
 
 | | Crystal structure of E. coli apoH-protein | | Descriptor: | Glycine cleavage system H protein | | Authors: | Fujiwara, K, Maita, N. | | Deposit date: | 2009-09-28 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
3A8K
 
 | | Crystal Structure of ETD97N-EHred complex | | Descriptor: | Aminomethyltransferase, Glycine cleavage system H protein | | Authors: | Okamura-Ikeda, K, Hosaka, H. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
2ZOG
 
 | | Crystal structure of mouse carnosinase CN2 complexed with ZN and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Cytosolic non-specific dipeptidase, ZINC ION | | Authors: | Unno, H, Yamashita, T, Okumura, N, Kusunoki, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for substrate recognition and hydrolysis by mouse carnosinase CN2.
J.Biol.Chem., 283, 2008
|
|
3AB9
 
 | | Crystal Structure of lipoylated E. coli H-protein (reduced form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycine cleavage system H protein | | Authors: | Okamura-Ikeda, K, Maita, N. | | Deposit date: | 2009-12-04 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
5DGR
 
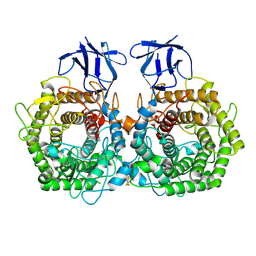 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | Authors: | Suzuki, K, Honda, Y, Fushinobu, S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
5DGQ
 
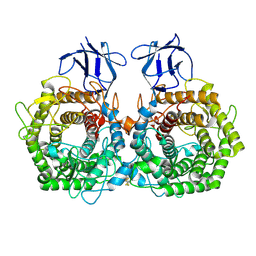 | |
2DEK
 
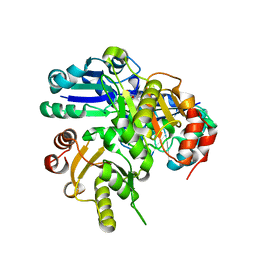 | |
2DQX
 
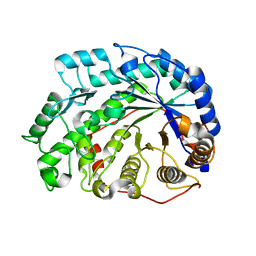 | | mutant beta-amylase (W55R) from soy bean | | Descriptor: | Beta-amylase | | Authors: | Ishikawa, K. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and structural analysis of enzyme sliding on a substrate: multiple attack in beta-amylase
Biochemistry, 46, 2007
|
|
2Z7Q
 
 | |
2Z7S
 
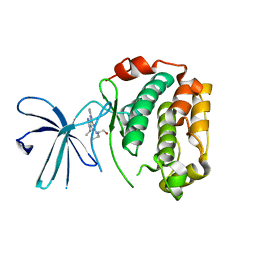 | | Crystal Structure of the N-terminal Kinase Domain of Human RSK1 bound to Purvalnol A | | Descriptor: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Ikuta, M, Munshi, S.K. | | Deposit date: | 2007-08-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: Implications for the design of RSK1 specific inhibitors.
Protein Sci., 16, 2007
|
|
2Z7R
 
 | |
3A8U
 
 | |
