7VNB
 
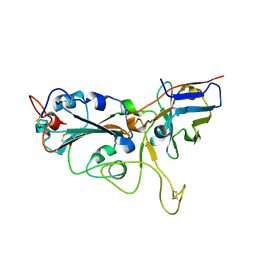 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNC
 
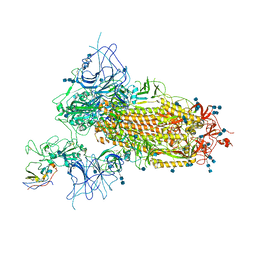 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7WHK
 
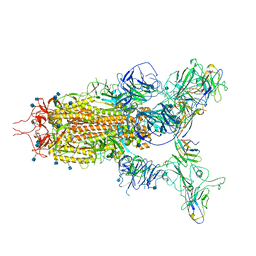 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHI
 
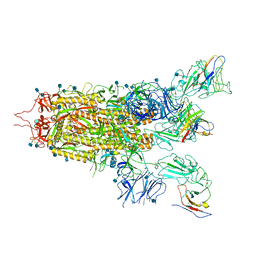 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHJ
 
 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7VK9
 
 | | Crystal structure of xCas9 P411T | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
7QEI
 
 | | Structure of human MTHFD2L in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Probable bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase 2 | | Authors: | Gustafsson, R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of Human MTHFD2L and Its Implications for the Development of Isoform-Selective Inhibitors.
Chemmedchem, 17, 2022
|
|
7VWV
 
 | |
7BOR
 
 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7CRD
 
 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRZ
 
 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7BU9
 
 | | Crystal Structure of Spindlin1-H3(K4me3-K9me2) complex | | Descriptor: | H3(K4me3-K9me2) peptide, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Molecular basis for histone H3 "K4me3-K9me3/2" methylation pattern readout by Spindlin1.
J.Biol.Chem., 295, 2020
|
|
7BQZ
 
 | |
4WZF
 
 | | Crystal structural basis for Rv0315, an immunostimulatory antigen and pseudo beta-1, 3-glucanase of Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanase, CALCIUM ION | | Authors: | Dong, W.Y, Fu, Z.F, Peng, G.Q. | | Deposit date: | 2014-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structural basis for Rv0315, an immunostimulatory antigen and inactive beta-1,3-glucanase of Mycobacterium tuberculosis.
Sci Rep, 5, 2015
|
|
8XMS
 
 | |
7AV4
 
 | | Dark state structure of the C432S mutant of Fatty Acid Photodecarboxylase (FAP) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Schlichting, I, Hartmann, E, Arnoux, P, Sorigue, D, Beisson, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7YZY
 
 | | pMMO structure from native membranes by cryoET and STA | | Descriptor: | Methane monooxygenase subunit C2, Particulate methane monooxygenase alpha subunit, Particulate methane monooxygenase beta subunit | | Authors: | Zhu, Y, Ni, T, Zhang, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and activity of particulate methane monooxygenase arrays in methanotrophs.
Nat Commun, 13, 2022
|
|
2OGD
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-527 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2007-01-05 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisphosphonates: Teaching Old Drugs with New Tricks
TO BE PUBLISHED
|
|
8AON
 
 | |
7ND4
 
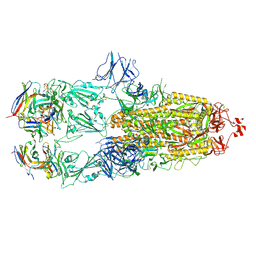 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-88 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
