5CGE
 
 | | Structure of Hydroxyethylthiazole Kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(2-methyl-1H-imidazole-1-yl)ethanol | | Descriptor: | 2-(2-methyl-1H-imidazol-1-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
7TUM
 
 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
2W89
 
 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
4J63
 
 | |
4J67
 
 | | Crystal structure of Ribonuclease A soaked in 50% 1,6-Hexanediol: One of twelve in MSCS set | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J60
 
 | |
4J64
 
 | | Crystal structure of Ribonuclease A soaked in 40% Dioxane: One of twelve in MSCS set | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J62
 
 | |
4J69
 
 | | Crystal structure of Ribonuclease A soaked in 50% S,R,S-bisfuranol: One of twelve in MSCS set | | Descriptor: | (3S,3aR,6aS)-hexahydrofuro[2,3-b]furan-3-ol, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J68
 
 | |
4J5Z
 
 | |
4J65
 
 | |
4J61
 
 | |
4J66
 
 | |
4J6A
 
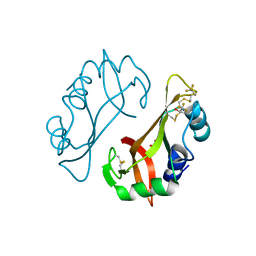 | | Crystal structure of Ribonuclease A soaked in 40% 2,2,2-Trifluoroethanol: One of twelve in MSCS set | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, TRIFLUOROETHANOL | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
6QY1
 
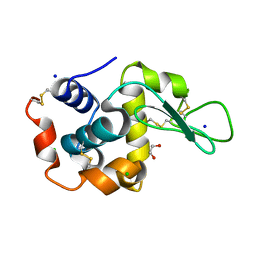 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 1500 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
6QXX
 
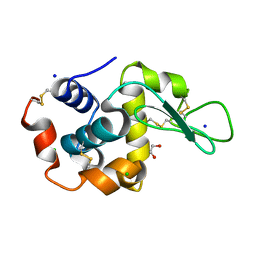 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 14793 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
6QXW
 
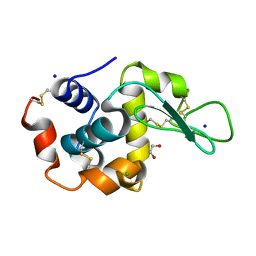 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 24344 patterns merged (3 chips) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
6QXY
 
 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 8813 patterns merged (1 chip) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
6QY2
 
 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 750 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
6QY5
 
 | | Pink beam serial crystallography: Lysozyme, 1 us exposure, 4448 patterns merged (1 chip) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
6QY0
 
 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 3000 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
