6ZJ9
 
 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
2XP8
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCC
 
 | | Crystal structure of PcrH from Pseudomonas aeruginosa | | Descriptor: | REGULATORY PROTEIN PCRH | | Authors: | Job, V, Mattei, P.-J, Lemaire, D, Attree, I, Dessen, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Chaperone Recognition of Type III Secretion System Minor Translocator Proteins.
J.Biol.Chem., 285, 2010
|
|
6ZL6
 
 | |
2XIL
 
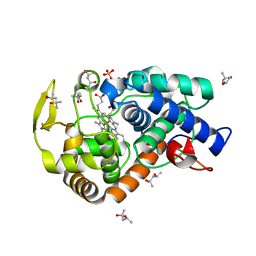 | | The structure of cytochrome c peroxidase Compound I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
6Z4U
 
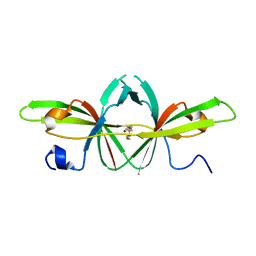 | |
2XI6
 
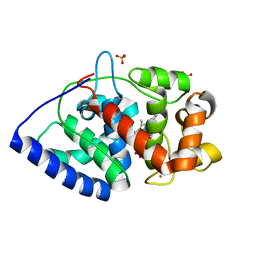 | | The structure of ascorbate peroxidase Compound I | | Descriptor: | ASCORBATE PEROXIDASE, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XIH
 
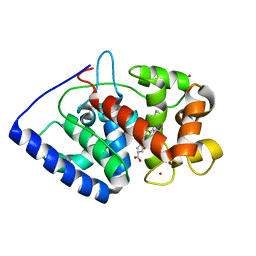 | | The structure of ascorbate peroxidase Compound III | | Descriptor: | ASCORBATE PEROXIDASE, OXYGEN MOLECULE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
6Z6U
 
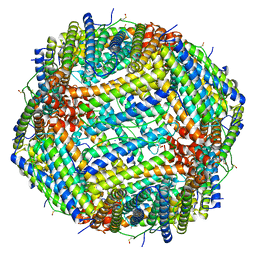 | | 1.25 A structure of human apoferritin obtained from Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-05-29 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.25 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6ZA1
 
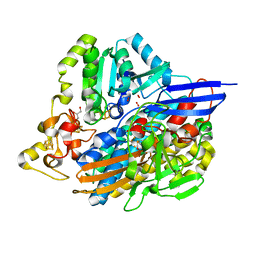 | | Structure of [NiFeSe] hydrogenase G491A variant from Desulfovibrio vulgaris Hildenborough pressurized with Oxygen gas - structure G491A-O2-hd | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, GLYCEROL, ... | | Authors: | Zacarias, S, Temporao, A, Carpentier, P, van der Linden, P, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
J.Biol.Inorg.Chem., 25, 2020
|
|
6Z9L
 
 | | Enterococcal PrgA | | Descriptor: | Poly-alanine peptide, PrgA, SULFATE ION | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
2XZ4
 
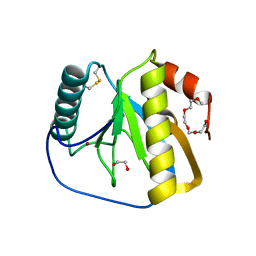 | | Crystal structure of the LFZ ectodomain of the peptidoglycan recognition protein LF | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COPPER (II) ION, ... | | Authors: | Basbous, N, Coste, F, Leone, P, Vincentelli, R, Royet, J, Kellenberger, C, Roussel, A. | | Deposit date: | 2010-11-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Drosophila Peptidoglycan-Recognition Protein Lf Interacts with Peptidoglycan-Recognition Protein Lc to Downregulate the Imd Pathway.
Embo Rep., 12, 2011
|
|
6ZCW
 
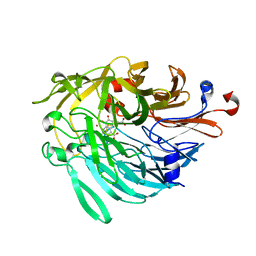 | |
6ZIG
 
 | |
2XC1
 
 | | Full-length Tailspike Protein Mutant Y108W of Bacteriophage P22 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, ... | | Authors: | Mueller, J.J, Seul, A, Seckler, R, Heinemann, U. | | Deposit date: | 2010-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6ZCO
 
 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | Descriptor: | Nucleoprotein | | Authors: | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
6ZCV
 
 | |
2XJ5
 
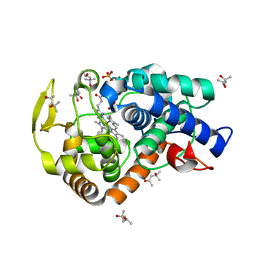 | | The structure of cytochrome c peroxidase Compound II | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XWA
 
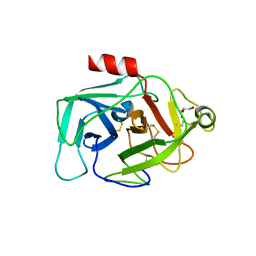 | | Crystal Structure of Complement Factor D Mutant R202A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
2XN5
 
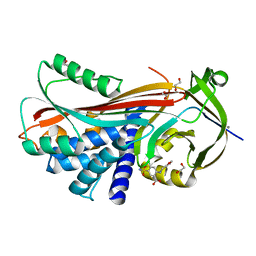 | | Crystal structure of thyroxine-binding globulin complexed with Furosemide | | Descriptor: | 1,2-ETHANEDIOL, 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Qi, X, Yan, Y, Wei, Z, Zhou, A. | | Deposit date: | 2010-07-30 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric Modulation of Hormone Release from Thyroxine and Corticosteroid Binding-Globulins.
J.Biol.Chem., 286, 2011
|
|
2XYK
 
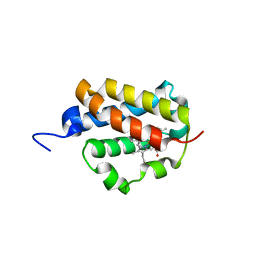 | | Group II 2-on-2 Hemoglobin from the Plant Pathogen Agrobacterium tumefaciens | | Descriptor: | 2-ON-2 HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, LaBarre, M, Richard, C, Wittenberg, J.B, Wittenberg, B.A, Guertin, M, Bolognesi, M. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of a Group II 2/2 Hemoglobin from the Plant Pathogen Agrobacterium Tumefaciens.
Biochim.Biophys.Acta, 1814, 2011
|
|
2XP9
 
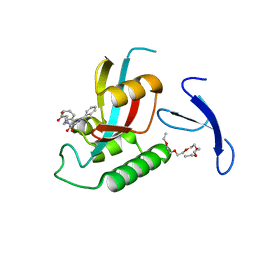 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[BENZYL(CARBOXYMETHYL)CARBAMOYL]-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6YWM
 
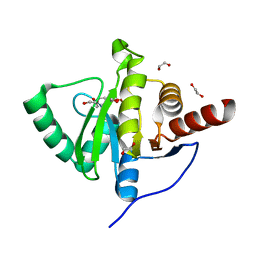 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YX5
 
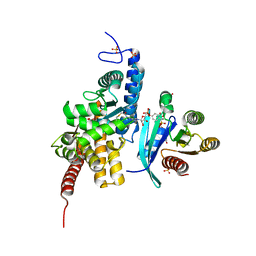 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
6Z9E
 
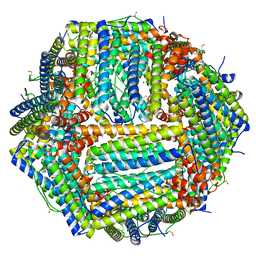 | | 1.55 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
