1SRK
 
 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
2JM3
 
 | | Solution structure of the THAP domain from C. elegans C-terminal binding protein (CtBP) | | Descriptor: | Hypothetical protein, ZINC ION | | Authors: | Liew, C.K, Crossley, M, Mackay, J.P, Nicholas, H.R. | | Deposit date: | 2006-09-21 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the THAP domain from Caenorhabditis elegans C-terminal binding protein (CtBP).
J.Mol.Biol., 366, 2007
|
|
2L6Z
 
 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID with FOG | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L6Y
 
 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7BGC
 
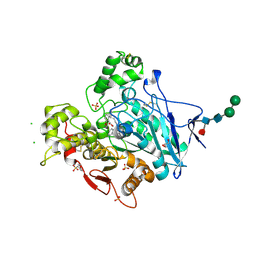 | | human butyrylcholinesterase in complex with a tacrine-methylanacardate hybrid inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Salerno, A, Bolognesi, M.L. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Drug Discovery of Multi-Target-Directed Ligands for Alzheimer's Disease.
J.Med.Chem., 64, 2021
|
|
4MXK
 
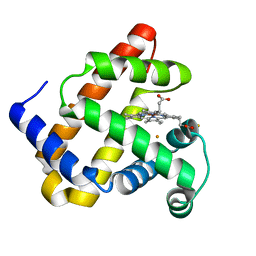 | | X-ray structure of Fe(II)-ZnPIXFeBMb1 | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Chakraborty, S, Lu, Y, Petrik, I. | | Deposit date: | 2013-09-26 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Spectroscopic and computational study of a nonheme iron nitrosyl center in a biosynthetic model of nitric oxide reductase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MXL
 
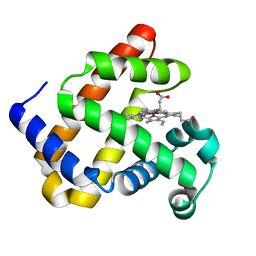 | | X-ray structure of ZnPFeBMb1 | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Chakraborty, S, Lu, Y, Petrik, I. | | Deposit date: | 2013-09-26 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Spectroscopic and computational study of a nonheme iron nitrosyl center in a biosynthetic model of nitric oxide reductase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5HAV
 
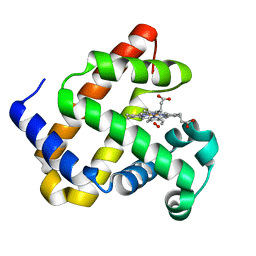 | |
2GG2
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 5-IMINO-4-(3-TRIFLUOROMETHYL-PHENYLAZO)-5H-PYRAZOL-3-YLAMINE, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG0
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-{(2S,3R)-3-AMINO-2-HYDROXY-3-[4-(TRIFLUOROMETHYL)PHENYL]PROPANOYL}ALANYLGLYCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GGB
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXYHEPTANOYL]-L-SERYL-L-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG8
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXY-3-(4-METHYLPHENYL)PROPANOYL]-D-ALANYL-D-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG7
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 3-(5-AMINO-3-IMINO-3H-PYRAZOL-4-YLAZO)-BENZOIC ACID, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG5
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 5-IMINO-4-(2-TRIFLUOROMETHYL-PHENYLAZO)-5H-PYRAZOL-3-YLAMINE, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG9
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXY-3-(4-ISOPROPYLPHENYL)PROPANOYL]-D-ALANYL-D-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG3
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 4-(4-FLUORO-PHENYLAZO)-5-IMINO-5H-PYRAZOL-3-YLAMINE, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
6ZJD
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
6ZJB
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Gp5A | | Descriptor: | GTP:AMP phosphotransferase AK3, mitochondrial, MAGNESIUM ION, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
6ZJE
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Ap5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
9FBV
 
 | |
5KCA
 
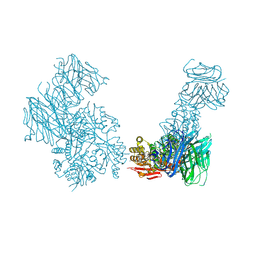 | | Crystal structure of the Cbln1 C1q domain trimer in complex with the amino-terminal domain (ATD) of iGluR Delta-2 (GluD2) | | Descriptor: | CALCIUM ION, Cerebellin-1,Cerebellin-1,Cerebellin-1,Glutamate receptor ionotropic, delta-2 | | Authors: | Elegheert, J, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC8
 
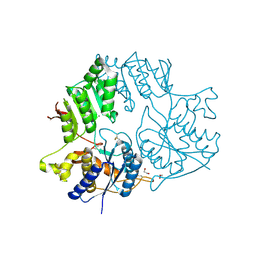 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-2 (GluD2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutamate receptor ionotropic, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC5
 
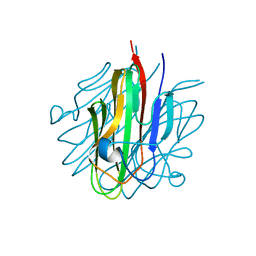 | | Crystal structure of the Cbln1 C1q domain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC9
 
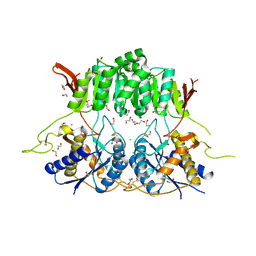 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-1 (GluD1) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC7
 
 | |
