1IC4
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC7
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1J2G
 
 | |
1J05
 
 | | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment | | Descriptor: | GLYCEROL, PHOSPHATE ION, anti-CEA mAb T84.66, ... | | Authors: | Kondo, H, Nishimura, Y, Shiroishi, M, Asano, R, Noro, N, Tsumoto, K, Kumagai, I. | | Deposit date: | 2002-11-01 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment
To be Published
|
|
1UA6
 
 | | Crystal structure of HYHEL-10 FV MUTANT SFSF complexed with HEN EGG WHITE LYSOZYME complex | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-02-28 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.Biol.Chem., 278, 2003
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
1IC5
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1UAC
 
 | | Crystal Structure of HYHEL-10 FV MUTANT SFSF Complexed with TURKEY WHITE LYSOZYME | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-03-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.BIOL.CHEM., 278, 2003
|
|
1VE9
 
 | | Porcine kidney D-amino acid oxidase | | Descriptor: | BENZOIC ACID, D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mizutani, H, Miyahara, I, Hirotsu, K, Nishina, Y, Shiga, K, Setoyama, C, Miura, R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0 A resolution.
J.Biochem., 120, 1996
|
|
5AUQ
 
 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5B40
 
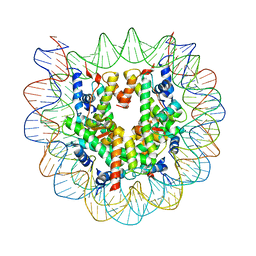 | | The nucleosome structure containing H2B-K120 and H4-K31 monoubiquitinations | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Machida, S, Sekine, S, Nishiyama, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Monoubiquitination of histones H2B and H4 changes the nucleosome stability without affecting the nucleosome structure
To Be Published
|
|
5AUN
 
 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUP
 
 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7FBH
 
 | | geranyl pyrophosphate C6-methyltransferase BezA | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BezA, ... | | Authors: | Tsutsumi, H, Moriwaki, Y, Terada, T, Shimizu, K, Katsuyama, Y, Ohnishi, Y. | | Deposit date: | 2021-07-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6-Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6KXE
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
5Y95
 
 | | Haddock model of mSIN3B PAH1 domain | | Descriptor: | Paired amphipathic helix protein Sin3b, propan-2-yl (3R,6S,9aS)-3-ethyl-8-(3-methylbutyl)-6-(2-methylsulfanylethyl)-4,7-bis(oxidanylidene)-9,9a-dihydro-6H-pyrazino[2,1-c][1,2,4]oxadiazine-1-carboxylate | | Authors: | Kurita, J, Hirao, Y, Nishimura, Y. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A mimetic of the mSin3-binding helix of NRSF/REST ameliorates abnormal pain behavior in chronic pain models.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6KXF
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KXD
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
3WP9
 
 | | Crystal structure of antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. | | Descriptor: | Ice-binding protein | | Authors: | Hanada, Y, Nishimiya, Y, Miura, A, Tsuda, S, Kondo, H. | | Deposit date: | 2014-01-10 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hyperactive antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. has a compound ice-binding site without repetitive sequences.
Febs J., 281, 2014
|
|
5XV8
 
 | |
6JK5
 
 | | Ca2+-dependent type II antifreeze protein (Ca2+-free form) | | Descriptor: | SULFATE ION, Type II antifreeze protein | | Authors: | Arai, T, Tsuda, S, Kondo, H, Nishimiya, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Calcium-Binding Generates the Semi-Clathrate Waters on a Type II Antifreeze Protein to Adsorb onto an Ice Crystal Surface.
Biomolecules, 9, 2019
|
|
6JK4
 
 | | Ca2+-dependent type II antifreeze protein | | Descriptor: | CALCIUM ION, Type II antifreeze protein | | Authors: | Arai, T, Tsuda, S, Kondo, H, Nishimiya, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Calcium-Binding Generates the Semi-Clathrate Waters on a Type II Antifreeze Protein to Adsorb onto an Ice Crystal Surface.
Biomolecules, 9, 2019
|
|
3VS8
 
 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
6JU6
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant | | Descriptor: | NITRATE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
